bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1315_orf3
Length=140
Score E
Sequences producing significant alignments: (Bits) Value
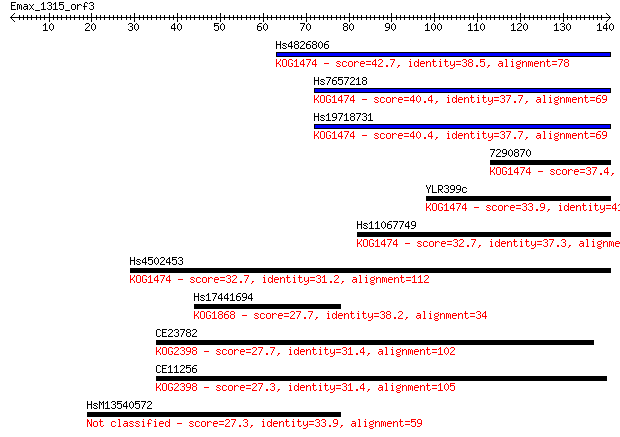
Hs4826806 42.7 2e-04
Hs7657218 40.4 0.001
Hs19718731 40.4 0.001
7290870 37.4 0.009
YLR399c 33.9 0.091
Hs11067749 32.7 0.23
Hs4502453 32.7 0.23
Hs17441694 27.7 7.9
CE23782 27.7 7.9
CE11256 27.3 8.3
HsM13540572 27.3 9.4
> Hs4826806
Length=801
Score = 42.7 bits (99), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/78 (38%), Positives = 41/78 (52%), Gaps = 6/78 (7%)
Query 63 GGVALLPNGPEPRAMYLSCGKMTTRSERGVMVRKPSKDLPAPISTPQPKTQRKVQLNEAL 122
G A P EP+A L R E G ++ P KDLP S Q ++ +K +L+E L
Sbjct 297 GSPASPPGSLEPKAARLP----PMRRESGRPIKPPRKDLPD--SQQQHQSSKKGKLSEQL 350
Query 123 RTCYEFVKELFTKKHAEY 140
+ C +KEL +KKHA Y
Sbjct 351 KHCNGILKELLSKKHAAY 368
> Hs7657218
Length=722
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 35/69 (50%), Gaps = 7/69 (10%)
Query 72 PEPRAMYLSCGKMTTRSERGVMVRKPSKDLPAPISTPQPKTQRKVQLNEALRTCYEFVKE 131
PEP+ K+ R E V+ P KD+P P P+ KV +E L+ C +KE
Sbjct 311 PEPKTT-----KLGQRRESSRPVKPPKKDVPDSQQHPAPEKSSKV--SEQLKCCSGILKE 363
Query 132 LFTKKHAEY 140
+F KKHA Y
Sbjct 364 MFAKKHAAY 372
> Hs19718731
Length=1362
Score = 40.4 bits (93), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 26/69 (37%), Positives = 35/69 (50%), Gaps = 7/69 (10%)
Query 72 PEPRAMYLSCGKMTTRSERGVMVRKPSKDLPAPISTPQPKTQRKVQLNEALRTCYEFVKE 131
PEP+ K+ R E V+ P KD+P P P+ KV +E L+ C +KE
Sbjct 311 PEPK-----TTKLGQRRESSRPVKPPKKDVPDSQQHPAPEKSSKV--SEQLKCCSGILKE 363
Query 132 LFTKKHAEY 140
+F KKHA Y
Sbjct 364 MFAKKHAAY 372
> 7290870
Length=1998
Score = 37.4 bits (85), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 14/28 (50%), Positives = 23/28 (82%), Gaps = 0/28 (0%)
Query 113 QRKVQLNEALRTCYEFVKELFTKKHAEY 140
+ K +L++AL++C E +KELF+KKH+ Y
Sbjct 472 KNKEKLSDALKSCNEILKELFSKKHSGY 499
> YLR399c
Length=686
Score = 33.9 bits (76), Expect = 0.091, Method: Compositional matrix adjust.
Identities = 18/43 (41%), Positives = 27/43 (62%), Gaps = 4/43 (9%)
Query 98 SKDLPAPISTPQPKTQRKVQLNEALRTCYEFVKELFTKKHAEY 140
SKD+ P + +PK++R L +A++ C +KEL KKHA Y
Sbjct 298 SKDI-YPYESKKPKSKR---LQQAMKFCQSVLKELMAKKHASY 336
> Hs11067749
Length=726
Score = 32.7 bits (73), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 32/60 (53%), Gaps = 3/60 (5%)
Query 82 GKMTTRSERGVM-VRKPSKDLPAPISTPQPKTQRKVQLNEALRTCYEFVKELFTKKHAEY 140
K+ R E G ++ P KDL PQ +K +L+E LR C ++E+ +KKHA Y
Sbjct 273 AKVVARRESGGRPIKPPKKDL-EDGEVPQ-HAGKKGKLSEHLRYCDSILREMLSKKHAAY 330
> Hs4502453
Length=947
Score = 32.7 bits (73), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 35/117 (29%), Positives = 52/117 (44%), Gaps = 8/117 (6%)
Query 29 PSIEEFEKTVAKKINKQRTASMGSEYSENGDTSTGGVALLPNG--PEPRAMYLSCGKMTT 86
PS+ F KT +N + AS+ S S+ T GV + P A+ S T
Sbjct 178 PSV--FPKTSISPLNVVQGASVNSS-SQTAAQVTKGVKRKADTTTPATSAVKASSEFSPT 234
Query 87 RSERGVMVRKPSKDLP---APISTPQPKTQRKVQLNEALRTCYEFVKELFTKKHAEY 140
+E+ V + +++P P S Q V++ E LR C E +KE+ KKH Y
Sbjct 235 FTEKSVALPPIKENMPKNVLPDSQQQYNVVETVKVTEQLRHCSEILKEMLAKKHFSY 291
> Hs17441694
Length=448
Score = 27.7 bits (60), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 13/37 (35%), Positives = 22/37 (59%), Gaps = 3/37 (8%)
Query 44 KQRTASMGSE---YSENGDTSTGGVALLPNGPEPRAM 77
K+R+ + GS +S+NG + + +LP+GP PR
Sbjct 286 KERSHANGSAVTMWSKNGPRNLSALEMLPDGPAPRTF 322
> CE23782
Length=968
Score = 27.7 bits (60), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 32/118 (27%), Positives = 52/118 (44%), Gaps = 17/118 (14%)
Query 35 EKTVAK--KINKQRTASMGSEYSENGDTSTGGVA-LLPNGPEPRAMYL-SCGKMTTRSER 90
E+TVA+ + K+R S+ EY + + S V+ + NG AM+L + G M +E
Sbjct 31 EETVAEVAQFVKERL-SVEDEYVKAINRSVNKVSHYIQNGSSIDAMWLLTKGTMELMAEI 89
Query 91 GVMVRKPSKDLPAPI------------STPQPKTQRKVQLNEALRTCYEFVKELFTKK 136
VM+ K +DL + QP+ V L + TC + KE + +
Sbjct 90 HVMLVKNLQDLSREVLKYKEDVNRTRKELKQPQVAEAVNLMQTTTTCLQKAKETYQHR 147
> CE11256
Length=951
Score = 27.3 bits (59), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 33/121 (27%), Positives = 53/121 (43%), Gaps = 17/121 (14%)
Query 35 EKTVAK--KINKQRTASMGSEYSENGDTSTGGVA-LLPNGPEPRAMYL-SCGKMTTRSER 90
E+TVA+ + K+R S+ EY + + S V+ + NG AM+L + G M +E
Sbjct 14 EETVAEVAQFVKER-LSVEDEYVKAINRSVNKVSHYIQNGSSIDAMWLLTKGTMELMAEI 72
Query 91 GVMVRKPSKDLPAPI------------STPQPKTQRKVQLNEALRTCYEFVKELFTKKHA 138
VM+ K +DL + QP+ V L + TC + KE + +
Sbjct 73 HVMLVKNLQDLSREVLKYKEDVNRTRKELKQPQVAEAVNLMQTTTTCLQKAKETYQHRCQ 132
Query 139 E 139
E
Sbjct 133 E 133
> HsM13540572
Length=127
Score = 27.3 bits (59), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 30/66 (45%), Gaps = 7/66 (10%)
Query 19 SEVEIPYPPRPSIEEFEKTVAKKINKQRTASMGSEYSENGDTSTGGVALLP-------NG 71
S V +P P PS+ + + + R S G + GDTS+ L+P +G
Sbjct 32 SAVILPQGPGPSLYLKQLCSSCQSQSTRRFSPGGSQLQCGDTSSSPTHLMPPPTPCPRSG 91
Query 72 PEPRAM 77
PEPR +
Sbjct 92 PEPRPL 97
Lambda K H
0.310 0.128 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1571531578
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40