bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
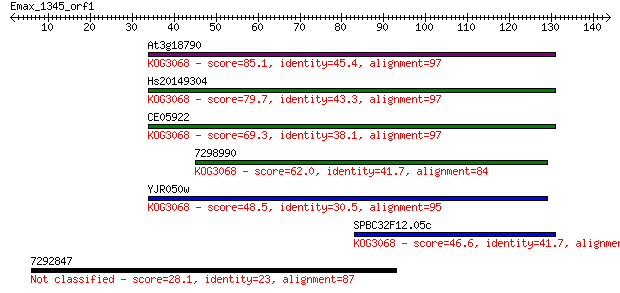
Query= Emax_1345_orf1
Length=143
Score E
Sequences producing significant alignments: (Bits) Value
At3g18790 85.1 4e-17
Hs20149304 79.7 2e-15
CE05922 69.3 2e-12
7298990 62.0 3e-10
YJR050w 48.5 4e-06
SPBC32F12.05c 46.6 1e-05
7292847 28.1 6.1
> At3g18790
Length=300
Score = 85.1 bits (209), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 44/97 (45%), Positives = 66/97 (68%), Gaps = 3/97 (3%)
Query 34 MARNAERANAVLNKWLAVKNAVIKGAKKNIQLGPCIPEECDDLDRATQWRQQLIREIGRG 93
MARN E+A ++LN+++ K + K K+ P + EC DL A +WRQQ++REIG
Sbjct 1 MARNEEKAQSMLNRFITQKESEKKKPKER---RPYLASECRDLAEADKWRQQILREIGSK 57
Query 94 ISLIQDSSLGDHRIRDLNDQINKKIKQKKRWEFRIKE 130
++ IQ+ LG+HR+RDLND+INK ++++ WE RI E
Sbjct 58 VAEIQNEGLGEHRLRDLNDEINKLLRERYHWERRIVE 94
> Hs20149304
Length=285
Score = 79.7 bits (195), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 42/97 (43%), Positives = 65/97 (67%), Gaps = 4/97 (4%)
Query 34 MARNAERANAVLNKWLAVKNAVIKGAKKNIQLGPCIPEECDDLDRATQWRQQLIREIGRG 93
MARNAE+A L ++ + A ++ K + P + EC +L +A +WR+Q+I EI +
Sbjct 1 MARNAEKAMTALARF---RQAQLEEGKVK-ERRPFLASECTELPKAEKWRRQIIGEISKK 56
Query 94 ISLIQDSSLGDHRIRDLNDQINKKIKQKKRWEFRIKE 130
++ IQ++ LG+ RIRDLND+INK +++K WE RIKE
Sbjct 57 VAQIQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKE 93
> CE05922
Length=267
Score = 69.3 bits (168), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 37/97 (38%), Positives = 61/97 (62%), Gaps = 3/97 (3%)
Query 34 MARNAERANAVLNKWLAVKNAVIKGAKKNIQLGPCIPEECDDLDRATQWRQQLIREIGRG 93
MARNAE+A L +W +K +G I P ++C +L A ++R++++R+ +
Sbjct 1 MARNAEKAMTALARWRRMKEEEERGP---IARRPHDVKDCRNLSDAERFRREIVRDASKK 57
Query 94 ISLIQDSSLGDHRIRDLNDQINKKIKQKKRWEFRIKE 130
I+ IQ+ LG+ ++RDLND++N+ IK K WE RI+E
Sbjct 58 ITAIQNPGLGEFKLRDLNDEVNRLIKLKHAWEQRIRE 94
> 7298990
Length=274
Score = 62.0 bits (149), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 35/88 (39%), Positives = 55/88 (62%), Gaps = 8/88 (9%)
Query 45 LNKWLAVKNAVIKGAKKNIQLGPCIPEECDDLDRATQWRQQLIREIGRGISLIQDSS--- 101
L +W A K V G K+ P + EC DL R ++R ++IR+I + ++ IQ+ +
Sbjct 10 LARWRAAKE-VESGEKERR---PYLASECHDLPRCEKFRLEIIRDISKKVAQIQNGTYFA 65
Query 102 -LGDHRIRDLNDQINKKIKQKKRWEFRI 128
LG+ RIRDLND+INK +++K+ WE +I
Sbjct 66 GLGEFRIRDLNDEINKLLREKRHWENQI 93
> YJR050w
Length=235
Score = 48.5 bits (114), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 29/96 (30%), Positives = 54/96 (56%), Gaps = 1/96 (1%)
Query 34 MARNAERANAVLNKWLAVKNAVIKGAKKNIQLG-PCIPEECDDLDRATQWRQQLIREIGR 92
M+RN ++AN+VL ++ + G K + P + + A +W++Q+ +EI +
Sbjct 1 MSRNVDKANSVLVRFQEQQAESAGGYKDYSRYQRPRNVSKVKSIKEANEWKRQVSKEIKQ 60
Query 93 GISLIQDSSLGDHRIRDLNDQINKKIKQKKRWEFRI 128
+ I D SL + +I +LND++N K+ KRW++ I
Sbjct 61 KSTRIYDPSLNEMQIAELNDELNNLFKEWKRWQWHI 96
> SPBC32F12.05c
Length=217
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 36/48 (75%), Gaps = 0/48 (0%)
Query 83 RQQLIREIGRGISLIQDSSLGDHRIRDLNDQINKKIKQKKRWEFRIKE 130
R ++++I R IS IQ ++L +++IRDLND IN+ +++K WE +I++
Sbjct 33 RASVVKDISRKISRIQSATLPEYQIRDLNDAINRLMREKHEWEVQIRD 80
> 7292847
Length=1547
Score = 28.1 bits (61), Expect = 6.1, Method: Composition-based stats.
Identities = 20/87 (22%), Positives = 38/87 (43%), Gaps = 1/87 (1%)
Query 6 TSTTAATATTTTTAAAAAAAAAAATAAKMARNAERANAVLNKWLAVKNAVIKGAKKNIQL 65
+S T + +T +A AA A ++ +L K +K V ++L
Sbjct 929 SSATDPSGSTAMSATVVAALNAVKNRKSQLSTQDKIQDLLQKKQELKGTVTPVGVNGVKL 988
Query 66 GPCIPEECDDLDRATQWRQQLIREIGR 92
P + D ++RA +++L+R+I R
Sbjct 989 NYGPPPK-DAIERACTMKEELLRKIER 1014
Lambda K H
0.318 0.128 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1639544670
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40