bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1368_orf2
Length=138
Score E
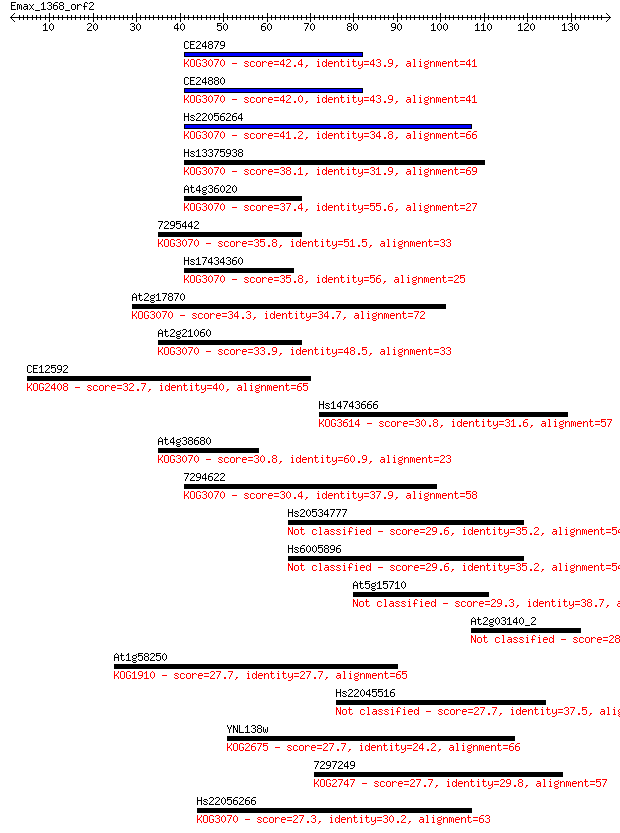
Sequences producing significant alignments: (Bits) Value
CE24879 42.4 3e-04
CE24880 42.0 4e-04
Hs22056264 41.2 6e-04
Hs13375938 38.1 0.005
At4g36020 37.4 0.009
7295442 35.8 0.022
Hs17434360 35.8 0.028
At2g17870 34.3 0.071
At2g21060 33.9 0.11
CE12592 32.7 0.20
Hs14743666 30.8 0.79
At4g38680 30.8 0.91
7294622 30.4 0.95
Hs20534777 29.6 1.7
Hs6005896 29.6 1.8
At5g15710 29.3 2.4
At2g03140_2 28.1 5.1
At1g58250 27.7 6.2
Hs22045516 27.7 7.2
YNL138w 27.7 7.4
7297249 27.7 7.6
Hs22056266 27.3 8.3
> CE24879
Length=227
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 18/41 (43%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 41 DIFVHQSYIQMQGFRALSVGDKVVFRIGVLPGKKAHQAVSV 81
D+FVHQS + MQGFR+L G++V + I K +A +V
Sbjct 77 DLFVHQSNLNMQGFRSLDEGERVSYYIQERSNGKGREAYAV 117
> CE24880
Length=196
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 18/41 (43%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 41 DIFVHQSYIQMQGFRALSVGDKVVFRIGVLPGKKAHQAVSV 81
D+FVHQS + MQGFR+L G++V + I K +A +V
Sbjct 46 DLFVHQSNLNMQGFRSLDEGERVSYYIQERSNGKGREAYAV 86
> Hs22056264
Length=157
Score = 41.2 bits (95), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 37/67 (55%), Gaps = 6/67 (8%)
Query 41 DIFVHQSYIQMQGFRALSVGDKVVFRIGVLPGKKAHQAV-SVQRTQESGKSVAATDSASK 99
D+FVHQS + M+GFR+L G+ V F KK+ + + S++ T G ++ K
Sbjct 82 DVFVHQSKLFMEGFRSLKEGEPVEFTF-----KKSSKGLESIRVTGPGGSPCLGSERRPK 136
Query 100 GDTEKQR 106
G T ++R
Sbjct 137 GKTLQKR 143
> Hs13375938
Length=209
Score = 38.1 bits (87), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 36/69 (52%), Gaps = 4/69 (5%)
Query 41 DIFVHQSYIQMQGFRALSVGDKVVFRIGVLPGKKAHQAVSVQRTQESGKSVAATDSASKG 100
D+FVHQS + M+GFR+L G+ V F K A S++ T G ++ KG
Sbjct 71 DVFVHQSKLHMEGFRSLKEGEAVEFTF----KKSAKGLESIRVTGPGGVFCIGSERRPKG 126
Query 101 DTEKQRSAE 109
+ ++R ++
Sbjct 127 KSMQKRRSK 135
> At4g36020
Length=299
Score = 37.4 bits (85), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 15/27 (55%), Positives = 21/27 (77%), Gaps = 0/27 (0%)
Query 41 DIFVHQSYIQMQGFRALSVGDKVVFRI 67
++FVHQS I +G+R+L+VGD V F I
Sbjct 35 ELFVHQSSIVSEGYRSLTVGDAVEFAI 61
> 7295442
Length=181
Score = 35.8 bits (81), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 17/34 (50%), Positives = 22/34 (64%), Gaps = 1/34 (2%)
Query 35 TPVSVG-DIFVHQSYIQMQGFRALSVGDKVVFRI 67
TP G ++FVHQS IQM GFR+L ++V F
Sbjct 42 TPNDGGQEVFVHQSVIQMSGFRSLGEQEEVEFEC 75
> Hs17434360
Length=181
Score = 35.8 bits (81), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 14/25 (56%), Positives = 19/25 (76%), Gaps = 0/25 (0%)
Query 41 DIFVHQSYIQMQGFRALSVGDKVVF 65
D+FVHQS + M+GFR+L G+ V F
Sbjct 130 DVFVHQSKLPMEGFRSLKEGEAVEF 154
> At2g17870
Length=299
Score = 34.3 bits (77), Expect = 0.071, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 41/73 (56%), Gaps = 4/73 (5%)
Query 29 KAHVVRTPVSVGD-IFVHQSYIQMQGFRALSVGDKVVFRIGVLPGKKAHQAVSVQRTQES 87
K + TP G+ +FVHQS I GFR+L++G+ V + I + K +A+ V T
Sbjct 20 KGYGFITPDDGGEELFVHQSSIVSDGFRSLTLGESVEYEIALGSDGKT-KAIEV--TAPG 76
Query 88 GKSVAATDSASKG 100
G S+ +++S+G
Sbjct 77 GGSLNKKENSSRG 89
> At2g21060
Length=201
Score = 33.9 bits (76), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 23/34 (67%), Gaps = 1/34 (2%)
Query 35 TPVSVGD-IFVHQSYIQMQGFRALSVGDKVVFRI 67
TP GD +FVHQS I+ +GFR+L+ + V F +
Sbjct 32 TPSDGGDDLFVHQSSIRSEGFRSLAAEESVEFDV 65
> CE12592
Length=773
Score = 32.7 bits (73), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 26/68 (38%), Positives = 38/68 (55%), Gaps = 7/68 (10%)
Query 5 TTTAGGVATQLKTL--DAKD-DIDTGVKAHVVRTPVSVGDIFVHQSYIQMQGFRALSVGD 61
T TA +A LKT+ D D D+ TG+ V+ TP++ G + S+I + FRAL GD
Sbjct 651 TFTAANLAA-LKTVYADPADIDLYTGL---VMETPLAGGQLGPTASWIIAEQFRALKTGD 706
Query 62 KVVFRIGV 69
+ + GV
Sbjct 707 RFYYENGV 714
> Hs14743666
Length=988
Score = 30.8 bits (68), Expect = 0.79, Method: Composition-based stats.
Identities = 18/57 (31%), Positives = 29/57 (50%), Gaps = 6/57 (10%)
Query 72 GKKAHQAVSVQRTQESGKSVAATDSASKGDTEKQRSAEEKPAIQQPAAPLFHTRQSL 128
G++A A+S Q S ++++ + K + SAEE P+AP HTR+S
Sbjct 885 GRRATIAISSQEGDNSERTLSNNITVPKIERANSYSAEE------PSAPYAHTRKSF 935
> At4g38680
Length=203
Score = 30.8 bits (68), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 14/24 (58%), Positives = 18/24 (75%), Gaps = 1/24 (4%)
Query 35 TPVSVGD-IFVHQSYIQMQGFRAL 57
TP GD +FVHQS I+ +GFR+L
Sbjct 28 TPDDGGDDLFVHQSSIRSEGFRSL 51
> 7294622
Length=340
Score = 30.4 bits (67), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 22/60 (36%), Positives = 32/60 (53%), Gaps = 4/60 (6%)
Query 41 DIFVHQSYIQMQGFRAL--SVGDKVVFRIGVLPGKKAHQAVSVQRTQESGKSVAATDSAS 98
D+FVHQS I + SVGD V V+ G+K ++A +V T SG+ V + A+
Sbjct 86 DVFVHQSAIARNNPKKAVRSVGDGEVVEFDVVIGEKGNEAANV--TGPSGEPVRGSQFAA 143
> Hs20534777
Length=571
Score = 29.6 bits (65), Expect = 1.7, Method: Composition-based stats.
Identities = 19/57 (33%), Positives = 25/57 (43%), Gaps = 4/57 (7%)
Query 65 FRIGVLPGKKAHQAVSVQRTQES---GKSVAATDSASKGDTEKQRSAEEKPAIQQPA 118
FR LP +AH+A+ QE G T G +E + EE+PA PA
Sbjct 499 FRASALPAAQAHEAMDCSILQEENGFGSRPQGTSPCPAGASE-EMEVEERPAGSTPA 554
> Hs6005896
Length=555
Score = 29.6 bits (65), Expect = 1.8, Method: Composition-based stats.
Identities = 19/57 (33%), Positives = 25/57 (43%), Gaps = 4/57 (7%)
Query 65 FRIGVLPGKKAHQAVSVQRTQES---GKSVAATDSASKGDTEKQRSAEEKPAIQQPA 118
FR LP +AH+A+ QE G T G +E + EE+PA PA
Sbjct 483 FRASALPAAQAHEAMDCSILQEENGFGSRPQGTSPCPAGASE-EMEVEERPAGSTPA 538
> At5g15710
Length=448
Score = 29.3 bits (64), Expect = 2.4, Method: Composition-based stats.
Identities = 12/31 (38%), Positives = 21/31 (67%), Gaps = 0/31 (0%)
Query 80 SVQRTQESGKSVAATDSASKGDTEKQRSAEE 110
SV+++ +SG S+A + SA GD E S+++
Sbjct 13 SVEKSLDSGNSLACSASAKNGDEESSTSSKQ 43
> At2g03140_2
Length=1298
Score = 28.1 bits (61), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 107 SAEEKPAIQQPAAPLFHTRQSLEEL 131
S + KPAIQ+P P F+ Q+ E L
Sbjct 417 SDQTKPAIQEPNQPNFNVSQAFEAL 441
> At1g58250
Length=2599
Score = 27.7 bits (60), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 38/65 (58%), Gaps = 1/65 (1%)
Query 25 DTGVKAHVVRTPVSVGDIFVHQSYIQMQGFRALSVGDKVVFRIGVLPGKKAHQAVSVQRT 84
+ G ++ +V + V V + ++ S+ ++Q ++ K +F+ + GKK ++ VSVQ +
Sbjct 543 EVGFRSKLVLS-VDVTGMGIYFSFKRVQSLIINALSFKALFKTLSVTGKKMNKTVSVQPS 601
Query 85 QESGK 89
+ SGK
Sbjct 602 KGSGK 606
> Hs22045516
Length=121
Score = 27.7 bits (60), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 26/48 (54%), Gaps = 1/48 (2%)
Query 76 HQAVSVQRTQESGKSVAATDSASKGDTEKQRSAEEKPAIQQPAAPLFH 123
H+ +V R QES AA D+AS+G T + +P++ A PL H
Sbjct 62 HRPAAVLRAQESDPGHAAADAASRG-TPPGGATSYRPSVNARAQPLRH 108
> YNL138w
Length=526
Score = 27.7 bits (60), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 16/66 (24%), Positives = 31/66 (46%), Gaps = 0/66 (0%)
Query 51 MQGFRALSVGDKVVFRIGVLPGKKAHQAVSVQRTQESGKSVAATDSASKGDTEKQRSAEE 110
MQG+ + + ++ L +Q +Q E+ K+ +DS + +T + SAE
Sbjct 8 MQGYNLVKLLKRLEEATARLEDVTIYQEGYIQNKLEASKNNKPSDSGADANTTNEPSAEN 67
Query 111 KPAIQQ 116
P ++Q
Sbjct 68 APEVEQ 73
> 7297249
Length=769
Score = 27.7 bits (60), Expect = 7.6, Method: Composition-based stats.
Identities = 17/57 (29%), Positives = 30/57 (52%), Gaps = 2/57 (3%)
Query 71 PGKKAHQAVSVQRTQESGKSVAATDSASKGDTEKQRSAEEKPAIQQPAAPLFHTRQS 127
P +K +++S++RT++ K TDS S GD E + K ++PA + +S
Sbjct 291 PMRKLTRSLSMRRTKQQPKQ--ETDSESDGDLEDDKIMISKSPAKKPAPSNLNASKS 345
> Hs22056266
Length=194
Score = 27.3 bits (59), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 33/64 (51%), Gaps = 6/64 (9%)
Query 44 VHQSYIQMQGFRALSVGDKVVFRIGVLPGKKAHQAV-SVQRTQESGKSVAATDSASKGDT 102
V +S + M+GFR+L G+ V F KK+ + + S++ T G ++ KG T
Sbjct 8 VSKSKLFMEGFRSLKEGEPVEFTF-----KKSSKGLESIRVTGPGGSPCLGSERRPKGKT 62
Query 103 EKQR 106
++R
Sbjct 63 LQKR 66
Lambda K H
0.312 0.126 0.343
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1498437086
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40