bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1421_orf1
Length=110
Score E
Sequences producing significant alignments: (Bits) Value
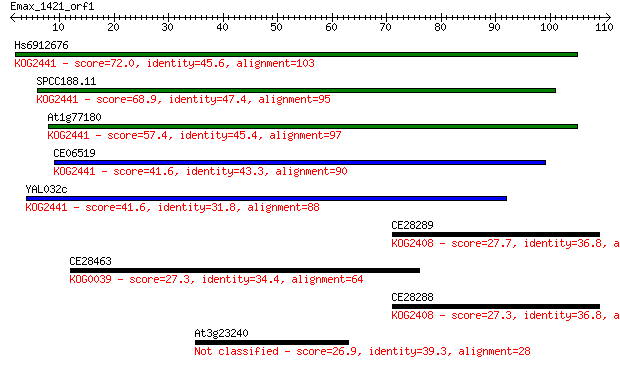
Hs6912676 72.0 3e-13
SPCC188.11 68.9 2e-12
At1g77180 57.4 6e-09
CE06519 41.6 3e-04
YAL032c 41.6 4e-04
CE28289 27.7 5.9
CE28463 27.3 6.4
CE28288 27.3 6.6
At3g23240 26.9 9.9
> Hs6912676
Length=536
Score = 72.0 bits (175), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 47/121 (38%), Positives = 66/121 (54%), Gaps = 24/121 (19%)
Query 2 IPQP----DSMFDARLFNQGGGTDSGYKGGEDDTYNLYDRPLFAQRGGA----GIYQFSR 53
+P P + +D RLFNQ G DSG+ GGED+ YN+YD+ A RGG IY+ S+
Sbjct 396 VPNPRTSNEVQYDQRLFNQSKGMDSGFAGGEDEIYNVYDQ---AWRGGKDMAQSIYRPSK 452
Query 54 DRFASSVGENSEL----------ASFAGADKTRYTRTGPVEFEKDVADPFGLDNLLSEAH 103
+ G++ E F+G+D+ + R GPV+FE+ DPFGLD L EA
Sbjct 453 NLDKDMYGDDLEARIKTNRFVPDKEFSGSDRRQRGREGPVQFEE---DPFGLDKFLEEAK 509
Query 104 K 104
+
Sbjct 510 Q 510
> SPCC188.11
Length=557
Score = 68.9 bits (167), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 45/112 (40%), Positives = 62/112 (55%), Gaps = 22/112 (19%)
Query 6 DSMFDARLFNQGGGTDSGYKGGEDDTYNLYDRPLFAQ------RGGAGIYQFSRDRFASS 59
D+M D+RLFNQ G SG++ ++D+YN+YD+P A R GA + SR AS+
Sbjct 444 DTMIDSRLFNQASGLGSGFQ--DEDSYNVYDKPWRAAPSSTLYRPGATL---SRQVDASA 498
Query 60 VGENSELAS-----------FAGADKTRYTRTGPVEFEKDVADPFGLDNLLS 100
E S F G+D+ +R GPV FEKD+ADPFG+D L+
Sbjct 499 ELERITSESRYDVLGNAHKKFKGSDEVVESRAGPVTFEKDIADPFGVDTFLN 550
> At1g77180
Length=613
Score = 57.4 bits (137), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 44/113 (38%), Positives = 60/113 (53%), Gaps = 18/113 (15%)
Query 8 MFDARLFNQGGGTDSGYKGGEDDTYNLYDRPLF-AQRGGAGIYQFSRDRFASSVG----- 61
M+D RLFNQ G DSG+ DD YNLYD+ LF AQ + +Y+ +D G
Sbjct 458 MYDQRLFNQDKGMDSGF--AADDQYNLYDKGLFTAQPTLSTLYKPKKDNDEEMYGNADEQ 515
Query 62 ----ENSEL----ASFAGA-DKTRYTRTGPVEFEK-DVADPFGLDNLLSEAHK 104
+N+E +F GA ++ R PVEFEK + DPFGL+ +S+ K
Sbjct 516 LDKIKNTERFKPDKAFTGASERVGSKRDRPVEFEKEEEQDPFGLEKWVSDLKK 568
> CE06519
Length=535
Score = 41.6 bits (96), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 39/112 (34%), Positives = 52/112 (46%), Gaps = 36/112 (32%)
Query 9 FDARLFNQGGGTDSGYKGGEDDTYNLYDRPLFAQRGGAGIYQF----------------- 51
FD RLF++ G DSG +DDTYN YD A RGG + Q
Sbjct 412 FDQRLFDKTQGLDSG--AMDDDTYNPYD---AAWRGGDSVQQHVYRPSKNLDNDVYGGDL 466
Query 52 -----SRDRFASSVGENSELASFAGADKTRYTRTGPVEFEKDVADPFGLDNL 98
++RF + G F+GA+ + +GPV+FEKD D FGL +L
Sbjct 467 DKIIEQKNRFVADKG-------FSGAEGSSRG-SGPVQFEKD-QDVFGLSSL 509
> YAL032c
Length=379
Score = 41.6 bits (96), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 28/88 (31%), Positives = 44/88 (50%), Gaps = 14/88 (15%)
Query 4 QPDSMFDARLFNQGGGTDSGYKGGEDDTYNLYDRPLFAQRGGAGIYQFSRDRFASSVGEN 63
QPD +D+R F +G ++ K ED +YD PLF Q+ IY+ + ++ +V
Sbjct 298 QPDLQYDSRFFTRGA--NASAKRHEDQ---VYDNPLFVQQDIESIYKTNYEKLDEAVNVK 352
Query 64 SELASFAGADKTRYTRTGPVEFEKDVAD 91
SE AS + GP++F K +D
Sbjct 353 SEGASGSH---------GPIQFTKAESD 371
> CE28289
Length=859
Score = 27.7 bits (60), Expect = 5.9, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 22/38 (57%), Gaps = 2/38 (5%)
Query 71 GADKTRYTRTGPVEFEKDVADPFGLDNLLSEAHKSPVE 108
GA YTR P ++ +V++P G +L ++A SP E
Sbjct 340 GASYRPYTRLLPTIYDNEVSEPVG--SLFTDARPSPRE 375
> CE28463
Length=1497
Score = 27.3 bits (59), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 32/69 (46%), Gaps = 9/69 (13%)
Query 12 RLFNQ-----GGGTDSGYKGGEDDTYNLYDRPLFAQRGGAGIYQFSRDRFASSVGENSEL 66
RLFN+ G D+ Y +D D P + G+ F+R + S+GE S L
Sbjct 900 RLFNEVLHYAGVSNDAKYLTYDDFNALFSDIP---DKQPVGL-PFNRKNYQPSIGETSSL 955
Query 67 ASFAGADKT 75
SFA D++
Sbjct 956 NSFAVVDRS 964
> CE28288
Length=1000
Score = 27.3 bits (59), Expect = 6.6, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 22/38 (57%), Gaps = 2/38 (5%)
Query 71 GADKTRYTRTGPVEFEKDVADPFGLDNLLSEAHKSPVE 108
GA YTR P ++ +V++P G +L ++A SP E
Sbjct 481 GASYRPYTRLLPTIYDNEVSEPVG--SLFTDARPSPRE 516
> At3g23240
Length=218
Score = 26.9 bits (58), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 11/28 (39%), Positives = 16/28 (57%), Gaps = 0/28 (0%)
Query 35 YDRPLFAQRGGAGIYQFSRDRFASSVGE 62
YD+ F+ RG + I FS +R S+ E
Sbjct 122 YDQAAFSMRGSSAILNFSAERVQESLSE 149
Lambda K H
0.314 0.136 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1195973986
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40