bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1447_orf1
Length=98
Score E
Sequences producing significant alignments: (Bits) Value
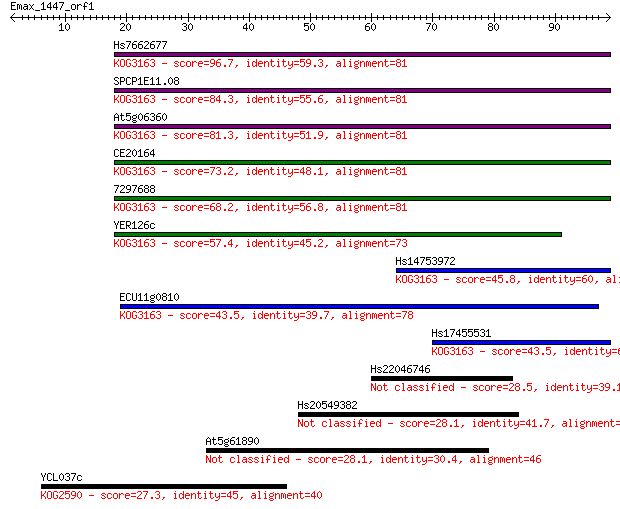
Hs7662677 96.7 9e-21
SPCP1E11.08 84.3 4e-17
At5g06360 81.3 4e-16
CE20164 73.2 1e-13
7297688 68.2 4e-12
YER126c 57.4 7e-09
Hs14753972 45.8 2e-05
ECU11g0810 43.5 9e-05
Hs17455531 43.5 1e-04
Hs22046746 28.5 3.4
Hs20549382 28.1 4.2
At5g61890 28.1 4.6
YCL037c 27.3 7.5
> Hs7662677
Length=260
Score = 96.7 bits (239), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 48/81 (59%), Positives = 63/81 (77%), Gaps = 0/81 (0%)
Query 18 ERKRKKAARTPHERSKTARKLRGIKAKLFNRRRFVEQAQMKTTIKHHEEKEGKQKEPETV 77
E+KRKK +R HERSK A+K+ G+KAKL++++R E+ QMK TIK HE++ KQK E
Sbjct 22 EKKRKKESREAHERSKKAKKMIGLKAKLYHKQRHAEKIQMKKTIKMHEKRNTKQKNDEKT 81
Query 78 PEGAVPSYLLDREGASRTKIL 98
P+GAVP+YLLDREG SR K+L
Sbjct 82 PQGAVPAYLLDREGQSRAKVL 102
> SPCP1E11.08
Length=260
Score = 84.3 bits (207), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 45/81 (55%), Positives = 57/81 (70%), Gaps = 0/81 (0%)
Query 18 ERKRKKAARTPHERSKTARKLRGIKAKLFNRRRFVEQAQMKTTIKHHEEKEGKQKEPETV 77
ERKRKKAAR H+ S A+K RGIKAKL+ +R E+ QMK TIK HEE+ Q+ +
Sbjct 22 ERKRKKAAREAHDASLYAQKTRGIKAKLYQEKRRKEKIQMKKTIKQHEERNATQRGSDAQ 81
Query 78 PEGAVPSYLLDREGASRTKIL 98
+GAVP+YLLDRE S+ K+L
Sbjct 82 TQGAVPTYLLDREQESQAKML 102
> At5g06360
Length=260
Score = 81.3 bits (199), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 42/81 (51%), Positives = 55/81 (67%), Gaps = 0/81 (0%)
Query 18 ERKRKKAARTPHERSKTARKLRGIKAKLFNRRRFVEQAQMKTTIKHHEEKEGKQKEPETV 77
ERKRKK AR H+ S A+K GIK K+ ++ + E+A MK T+K HEE ++K E V
Sbjct 22 ERKRKKEAREVHKHSTMAQKSLGIKGKMIAKKNYAEKALMKKTLKMHEESSSRRKADENV 81
Query 78 PEGAVPSYLLDREGASRTKIL 98
EGAVP+YLLDRE +R K+L
Sbjct 82 QEGAVPAYLLDREDTTRAKVL 102
> CE20164
Length=259
Score = 73.2 bits (178), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 39/82 (47%), Positives = 58/82 (70%), Gaps = 3/82 (3%)
Query 18 ERKRKKAARTPHERSKTARKLRGIKAKLFNRRRFVEQAQMKTTIKHHEEKEGKQKEPETV 77
ER+RKK AR H+RS+ A+ LRG KAKL++++R+ E+ +M+ +K HEEK+ QK
Sbjct 22 ERQRKKLARAAHDRSQMAKTLRGHKAKLYHKKRYSEKVEMRKLLKQHEEKD--QKNTVEQ 79
Query 78 PE-GAVPSYLLDREGASRTKIL 98
P+ GAVP+YLLDR+ + +L
Sbjct 80 PDKGAVPAYLLDRQQQTSGTVL 101
> 7297688
Length=253
Score = 68.2 bits (165), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 46/81 (56%), Positives = 59/81 (72%), Gaps = 1/81 (1%)
Query 18 ERKRKKAARTPHERSKTARKLRGIKAKLFNRRRFVEQAQMKTTIKHHEEKEGKQKEPETV 77
ERKRKK AR P +R++ ARKLRGIKAKLFN+ R E+ Q+K I+ HEEK+ +K+ E V
Sbjct 16 ERKRKKEARLPKDRARKARKLRGIKAKLFNKERRNEKIQIKKKIQAHEEKK-VKKQEEKV 74
Query 78 PEGAVPSYLLDREGASRTKIL 98
+GA+P YLLDR S K+L
Sbjct 75 EDGALPHYLLDRGIQSSAKVL 95
> YER126c
Length=261
Score = 57.4 bits (137), Expect = 7e-09, Method: Compositional matrix adjust.
Identities = 33/74 (44%), Positives = 47/74 (63%), Gaps = 1/74 (1%)
Query 18 ERKRKKAARTPHERSKTARKLRGIKAKLFNRRRFVEQAQMKTTIKHHEEKEGK-QKEPET 76
ERKRK+ AR H+ S+ A+KL G K K F ++R+ E+ M+ IK HE+ + K +P
Sbjct 22 ERKRKREARESHKISERAQKLTGWKGKQFAKKRYAEKVSMRKKIKAHEQSKVKGSSKPLD 81
Query 77 VPEGAVPSYLLDRE 90
A+P+YLLDRE
Sbjct 82 TDGDALPTYLLDRE 95
> Hs14753972
Length=194
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 21/35 (60%), Positives = 26/35 (74%), Gaps = 0/35 (0%)
Query 64 HEEKEGKQKEPETVPEGAVPSYLLDREGASRTKIL 98
HE++ KQK E P+GAVP+YLLDRE SR K+L
Sbjct 2 HEKRNIKQKNDEKTPQGAVPAYLLDREEQSRAKVL 36
> ECU11g0810
Length=258
Score = 43.5 bits (101), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 31/79 (39%), Positives = 47/79 (59%), Gaps = 4/79 (5%)
Query 19 RKRKKAARTPHERSKTARKLRGIKAKLFNRRRFVEQAQMKTTIKHHEEKEGKQKEPE-TV 77
RK KK R +KTA++L G+KAKLF +++ + Q+K +K KE K+K E +V
Sbjct 23 RKYKKERRNEKLVAKTAKRLFGLKAKLFAKKQHAAKIQLKKDLKM---KESKKKGMEVSV 79
Query 78 PEGAVPSYLLDREGASRTK 96
+ +P +LLDRE R +
Sbjct 80 EDNPLPHFLLDREMQERAR 98
> Hs17455531
Length=188
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 19/29 (65%), Positives = 23/29 (79%), Gaps = 0/29 (0%)
Query 70 KQKEPETVPEGAVPSYLLDREGASRTKIL 98
K+K E P+GAVP+YLLDREG SR K+L
Sbjct 8 KKKNAEKTPQGAVPAYLLDREGQSRAKVL 36
> Hs22046746
Length=223
Score = 28.5 bits (62), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 9/23 (39%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 60 TIKHHEEKEGKQKEPETVPEGAV 82
+KHH E ++ EP+T P+G +
Sbjct 19 VLKHHNEAPDEESEPDTCPQGGL 41
> Hs20549382
Length=435
Score = 28.1 bits (61), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 20/36 (55%), Gaps = 1/36 (2%)
Query 48 RRRFVEQAQMKTTIKHHEEKEGKQKEPETVPEGAVP 83
+R F EQ + KH +E+ G K+P PEGA P
Sbjct 29 QRLFPEQRACSSRGKHGDER-GAPKDPPAAPEGAFP 63
> At5g61890
Length=248
Score = 28.1 bits (61), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 14/46 (30%), Positives = 25/46 (54%), Gaps = 4/46 (8%)
Query 33 KTARKLRGIKAKLFNRRRFVEQAQMKTTIKHHEEKEGKQKEPETVP 78
+ A K +G KAKL F E+ Q+ + ++ + Q EP+++P
Sbjct 131 EAALKFKGSKAKL----NFPERVQLGSNSTYYSSNQIPQMEPQSIP 172
> YCL037c
Length=466
Score = 27.3 bits (59), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 22/44 (50%), Gaps = 4/44 (9%)
Query 6 LLPTFFPFSNTA----ERKRKKAARTPHERSKTARKLRGIKAKL 45
L PT P S + + RKK RTP +S TA K IKA +
Sbjct 76 LAPTEIPVSTISIEDLDATRKKKNRTPTPKSSTATKWVPIKASI 119
Lambda K H
0.315 0.129 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1194657780
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40