bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1475_orf1
Length=144
Score E
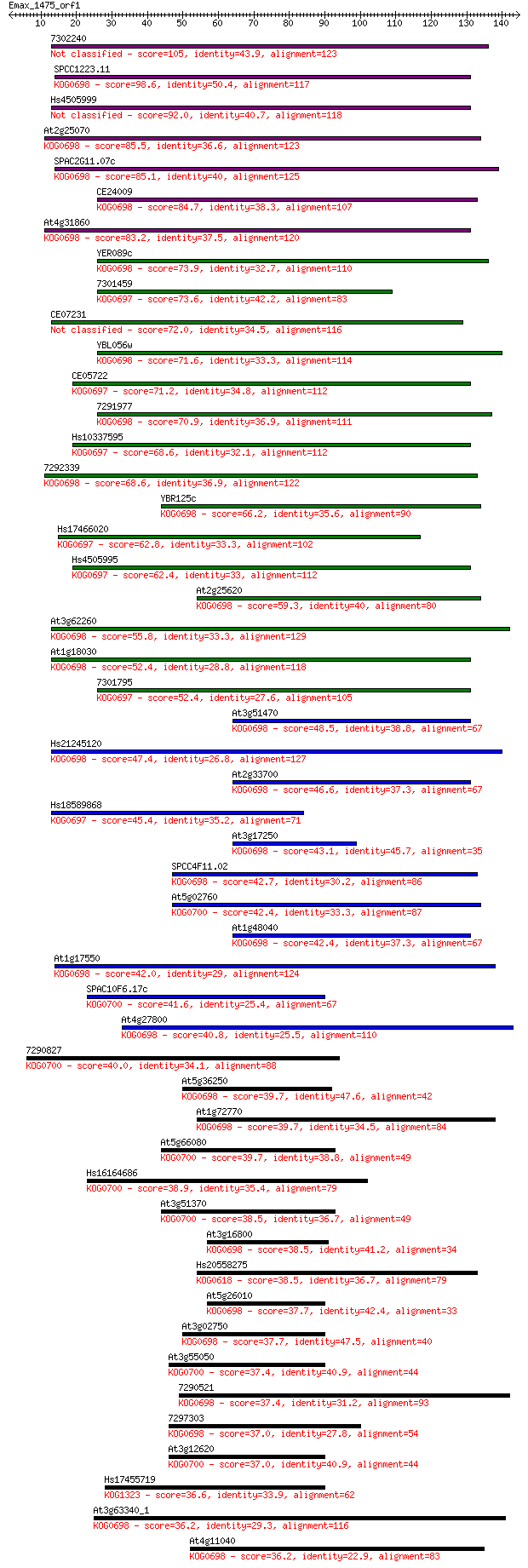
Sequences producing significant alignments: (Bits) Value
7302240 105 2e-23
SPCC1223.11 98.6 4e-21
Hs4505999 92.0 3e-19
At2g25070 85.5 3e-17
SPAC2G11.07c 85.1 4e-17
CE24009 84.7 5e-17
At4g31860 83.2 2e-16
YER089c 73.9 8e-14
7301459 73.6 1e-13
CE07231 72.0 4e-13
YBL056w 71.6 5e-13
CE05722 71.2 5e-13
7291977 70.9 7e-13
Hs10337595 68.6 3e-12
7292339 68.6 4e-12
YBR125c 66.2 2e-11
Hs17466020 62.8 2e-10
Hs4505995 62.4 3e-10
At2g25620 59.3 3e-09
At3g62260 55.8 3e-08
At1g18030 52.4 3e-07
7301795 52.4 3e-07
At3g51470 48.5 4e-06
Hs21245120 47.4 8e-06
At2g33700 46.6 1e-05
Hs18589868 45.4 3e-05
At3g17250 43.1 2e-04
SPCC4F11.02 42.7 3e-04
At5g02760 42.4 3e-04
At1g48040 42.4 3e-04
At1g17550 42.0 4e-04
SPAC10F6.17c 41.6 5e-04
At4g27800 40.8 8e-04
7290827 40.0 0.001
At5g36250 39.7 0.002
At1g72770 39.7 0.002
At5g66080 39.7 0.002
Hs16164686 38.9 0.004
At3g51370 38.5 0.004
At3g16800 38.5 0.004
Hs20558275 38.5 0.005
At5g26010 37.7 0.007
At3g02750 37.7 0.008
At3g55050 37.4 0.009
7290521 37.4 0.010
7297303 37.0 0.012
At3g12620 37.0 0.014
Hs17455719 36.6 0.016
At3g63340_1 36.2 0.020
At4g11040 36.2 0.020
> 7302240
Length=662
Score = 105 bits (263), Expect = 2e-23, Method: Composition-based stats.
Identities = 54/123 (43%), Positives = 76/123 (61%), Gaps = 1/123 (0%)
Query 13 GGRISRNGSCKWELELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIG 72
GGR++ +G L L + G YK + +LP E+Q++S PD+ + PE DEF+V+
Sbjct 446 GGRVTLDGRVNGGLNLSRALGDHAYKTNVTLPAEEQMISALPDIKKLIITPE-DEFMVLA 504
Query 73 CDGIWELLSSAEVVEFVRRRIEHTPDLCQILKELFDSLISPNPALFEYGCDNMTAFIVDL 132
CDGIW +SS EVVEFVR R++ L I +ELFD+ ++PN GCDNMTA IV
Sbjct 505 CDGIWNYMSSEEVVEFVRCRLKDNKKLSTICEELFDNCLAPNTMGDGTGCDNMTAVIVQF 564
Query 133 QAE 135
+ +
Sbjct 565 KKK 567
> SPCC1223.11
Length=370
Score = 98.6 bits (244), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 59/117 (50%), Positives = 70/117 (59%), Gaps = 7/117 (5%)
Query 14 GRISRNGSCKWELELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIGC 73
GR+ NGS L L + G YK D SLPPEKQI++ PDVV N DP+ DEFL++ C
Sbjct 180 GRV--NGS----LALSRAIGDFEYKKDSSLPPEKQIVTAFPDVVIHNIDPD-DEFLILAC 232
Query 74 DGIWELLSSAEVVEFVRRRIEHTPDLCQILKELFDSLISPNPALFEYGCDNMTAFIV 130
DGIW+ SS +VVEFVRR I L I + L D I+ N GCDNMT IV
Sbjct 233 DGIWDCKSSQQVVEFVRRGIVARQSLEVICENLMDRCIASNSESCGIGCDNMTICIV 289
> Hs4505999
Length=546
Score = 92.0 bits (227), Expect = 3e-19, Method: Compositional matrix adjust.
Identities = 48/126 (38%), Positives = 75/126 (59%), Gaps = 11/126 (8%)
Query 13 GGRISRNGSCKWELELVKGFGRLGYKADDSLPPEKQILSGCPD--VVTVNRDPEKDEFLV 70
GG+++ +G L L + G YK + +LPPE+Q++S PD V+T+ D EF+V
Sbjct 381 GGKVTMDGRVNGGLNLSRAIGDHFYKRNKNLPPEEQMISALPDIKVLTLTDD---HEFMV 437
Query 71 IGCDGIWELLSSAEVVEFVRRRIEHTPD------LCQILKELFDSLISPNPALFEYGCDN 124
I CDGIW ++SS EVV+F++ +I + L I++EL D ++P+ + GCDN
Sbjct 438 IACDGIWNVMSSQEVVDFIQSKISQRDENGELRLLSSIVEELLDQCLAPDTSGDGTGCDN 497
Query 125 MTAFIV 130
MT I+
Sbjct 498 MTCIII 503
> At2g25070
Length=355
Score = 85.5 bits (210), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 45/123 (36%), Positives = 75/123 (60%), Gaps = 8/123 (6%)
Query 11 IRGGRISRNGSCKWELELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLV 70
I GRI NGS L L + G + +K + LP EKQ+++ PD+ T++ + D+FLV
Sbjct 216 IHAGRI--NGS----LNLTRAIGDMEFKQNKFLPSEKQMVTADPDINTIDL-CDDDDFLV 268
Query 71 IGCDGIWELLSSAEVVEFVRRRIEHTPDLCQILKELFDSLISPNPALFEYGCDNMTAFIV 130
+ CDGIW+ +SS E+V+F+ +++ L + +++ D ++P+ A E GCDNMT +V
Sbjct 269 VACDGIWDCMSSQELVDFIHEQLKSETKLSTVCEKVVDRCLAPDTATGE-GCDNMTIILV 327
Query 131 DLQ 133
+
Sbjct 328 QFK 330
> SPAC2G11.07c
Length=414
Score = 85.1 bits (209), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 50/125 (40%), Positives = 72/125 (57%), Gaps = 8/125 (6%)
Query 14 GRISRNGSCKWELELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIGC 73
GR++ N L L + G +K + +L PEKQI++ PDVV V+ + DEF+V+ C
Sbjct 178 GRVNGN------LALSRAIGDFEFK-NSNLEPEKQIVTALPDVV-VHEITDDDEFVVLAC 229
Query 74 DGIWELLSSAEVVEFVRRRIEHTPDLCQILKELFDSLISPNPALFEYGCDNMTAFIVDLQ 133
DGIW+ +S +V+EFVRR I L +I + L D+ I+ + GCDNMT IV L
Sbjct 230 DGIWDCKTSQQVIEFVRRGIVAGTSLEKIAENLMDNCIASDTETTGLGCDNMTVCIVALL 289
Query 134 AEKRR 138
E +
Sbjct 290 QENDK 294
> CE24009
Length=356
Score = 84.7 bits (208), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 41/107 (38%), Positives = 65/107 (60%), Gaps = 1/107 (0%)
Query 26 LELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEV 85
L L + G +K D+ P E+QI++ PDV+T P+ EF+V+ CDGIW+++++ EV
Sbjct 181 LALSRALGDFAFKNCDTKPAEEQIVTAFPDVITDKLTPDH-EFIVLACDGIWDVMTNQEV 239
Query 86 VEFVRRRIEHTPDLCQILKELFDSLISPNPALFEYGCDNMTAFIVDL 132
V+FVR ++ D I +EL ++P+ + GCDNMT +V L
Sbjct 240 VDFVREKLAEKRDPQSICEELLTRCLAPDCQMGGLGCDNMTVVLVGL 286
> At4g31860
Length=357
Score = 83.2 bits (204), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 45/120 (37%), Positives = 72/120 (60%), Gaps = 8/120 (6%)
Query 11 IRGGRISRNGSCKWELELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLV 70
I GR+ NGS L L + G + +K + LP EKQI++ PDV TV + D+FLV
Sbjct 216 IHAGRV--NGS----LNLSRAIGDMEFKQNKFLPSEKQIVTASPDVNTVEL-CDDDDFLV 268
Query 71 IGCDGIWELLSSAEVVEFVRRRIEHTPDLCQILKELFDSLISPNPALFEYGCDNMTAFIV 130
+ CDGIW+ ++S ++V+F+ ++ L + +++ D ++PN + E GCDNMT +V
Sbjct 269 LACDGIWDCMTSQQLVDFIHEQLNSETKLSVVCEKVLDRCLAPNTSGGE-GCDNMTMILV 327
> YER089c
Length=464
Score = 73.9 bits (180), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 36/110 (32%), Positives = 61/110 (55%), Gaps = 0/110 (0%)
Query 26 LELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEV 85
L L + G +K++ L PE+QI++ PD++ + D ++DEF+++ CDGIW+ L+S +
Sbjct 186 LALSRAIGDFEFKSNPKLGPEEQIVTCVPDILEHSLDYDRDEFVILACDGIWDCLTSQDC 245
Query 86 VEFVRRRIEHTPDLCQILKELFDSLISPNPALFEYGCDNMTAFIVDLQAE 135
V+ V + L +I + D +P GCDNM+ +V L E
Sbjct 246 VDLVHLGLREGKTLNEISSRIIDVCCAPTTEGTGIGCDNMSIVVVALLKE 295
> 7301459
Length=367
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 35/83 (42%), Positives = 53/83 (63%), Gaps = 1/83 (1%)
Query 26 LELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEV 85
L + + FG +K D S P Q++S PD++ NR E DEF+V+ CDGIW++++S+EV
Sbjct 180 LAVSRAFGDYDFKNDGSKSPVDQMVSPEPDIIVCNRS-EHDEFIVVACDGIWDVMTSSEV 238
Query 86 VEFVRRRIEHTPDLCQILKELFD 108
EF+R R+ T DL I+ + D
Sbjct 239 CEFIRSRLLVTYDLPMIVNSVLD 261
> CE07231
Length=491
Score = 72.0 bits (175), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 40/116 (34%), Positives = 61/116 (52%), Gaps = 2/116 (1%)
Query 13 GGRISRNGSCKWELELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIG 72
GG+I +G L L + FG YK + L ++Q+++ PDV PE DEF+V+
Sbjct 369 GGQI-EDGRVNGGLNLSRAFGDHAYKKNQELGLKEQMITALPDVKIEALTPE-DEFIVVA 426
Query 73 CDGIWELLSSAEVVEFVRRRIEHTPDLCQILKELFDSLISPNPALFEYGCDNMTAF 128
CDGIW + S +VV+FVR + ++ L D+ ++ + GCDNMT
Sbjct 427 CDGIWNSMESQQVVDFVRDLLAKGSSCAEVCDALCDACLADSTDGDGTGCDNMTVI 482
> YBL056w
Length=468
Score = 71.6 bits (174), Expect = 5e-13, Method: Composition-based stats.
Identities = 38/115 (33%), Positives = 60/115 (52%), Gaps = 1/115 (0%)
Query 26 LELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEV 85
L L + G +K++ L P +Q+++ PD++ N + ++DEF+++ CDGIW+ L+S E
Sbjct 186 LALSRAIGDFEFKSNTKLGPHEQVVTCVPDIICHNLNYDEDEFVILACDGIWDCLTSQEC 245
Query 86 VEFVRRRIEH-TPDLCQILKELFDSLISPNPALFEYGCDNMTAFIVDLQAEKRRQ 139
V+ V I L I + D SP GCDNM+ IV L E +
Sbjct 246 VDLVHYGISQGNMTLSDISSRIVDVCCSPTTEGSGIGCDNMSISIVALLKENESE 300
> CE05722
Length=468
Score = 71.2 bits (173), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 39/112 (34%), Positives = 64/112 (57%), Gaps = 10/112 (8%)
Query 19 NGSCKWELELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIGCDGIWE 78
NGS L + + FG YK D LP ++Q++S PDV R+ E D+F+V+ CDGI++
Sbjct 278 NGS----LAVSRAFGDYEYKDDPRLPADQQLVSPEPDVYIRERNLENDQFMVVACDGIYD 333
Query 79 LLSSAEVVEFVRRRIEHTPDLCQILKELFDSLISPNPALFEYGCDNMTAFIV 130
++++ E+ EFV+ R+ DL ++ ++ D L + DNMT +V
Sbjct 334 VMTNEELAEFVKDRLSVHSDLREVCDDVLDE------CLVKGSRDNMTMVVV 379
> 7291977
Length=352
Score = 70.9 bits (172), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 41/111 (36%), Positives = 63/111 (56%), Gaps = 1/111 (0%)
Query 26 LELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEV 85
L L + G YK + PE+QI++ PDV ++ E EF+++ CDGIW+++S+ EV
Sbjct 181 LALSRALGDFIYKKNLLKTPEEQIVTAYPDVEVLDI-TEDLEFVLLACDGIWDVMSNFEV 239
Query 86 VEFVRRRIEHTPDLCQILKELFDSLISPNPALFEYGCDNMTAFIVDLQAEK 136
+FV +RI + I +EL +S +SP+ G DNMT +V L K
Sbjct 240 CQFVHKRIRDGMEPELICEELMNSCLSPDGHTGNVGGDNMTVILVCLLHNK 290
> Hs10337595
Length=382
Score = 68.6 bits (166), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 36/112 (32%), Positives = 63/112 (56%), Gaps = 10/112 (8%)
Query 19 NGSCKWELELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIGCDGIWE 78
NGS L + + G YK P +Q++S P+V + R E D+F+++ CDGIW+
Sbjct 188 NGS----LAVSRALGDFDYKCVHGKGPTEQLVSPEPEVHDIERSEEDDQFIILACDGIWD 243
Query 79 LLSSAEVVEFVRRRIEHTPDLCQILKELFDSLISPNPALFEYGCDNMTAFIV 130
++ + E+ +FVR R+E T DL ++ E+ D+ L++ DNM+ ++
Sbjct 244 VMGNEELCDFVRSRLEVTDDLEKVCNEVVDT------CLYKGSRDNMSVILI 289
> 7292339
Length=371
Score = 68.6 bits (166), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 45/123 (36%), Positives = 67/123 (54%), Gaps = 4/123 (3%)
Query 11 IRGGRISRNGSCKWELELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKD-EFL 69
I+GG L L + G +K ++ PE QI++ PDV T R D EF+
Sbjct 165 IQGGGWVEFNRVNGNLALSRALGDYVFKHENK-KPEDQIVTAFPDVET--RKIMDDWEFI 221
Query 70 VIGCDGIWELLSSAEVVEFVRRRIEHTPDLCQILKELFDSLISPNPALFEYGCDNMTAFI 129
V+ CDGIW+++S+AEV+EF R RI +I +EL + ++P+ + G DNMT +
Sbjct 222 VLACDGIWDVMSNAEVLEFCRTRIGMGMFPEEICEELMNHCLAPDCQMGGLGGDNMTVVL 281
Query 130 VDL 132
V L
Sbjct 282 VCL 284
> YBR125c
Length=393
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 32/90 (35%), Positives = 52/90 (57%), Gaps = 0/90 (0%)
Query 44 PPEKQILSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEVVEFVRRRIEHTPDLCQIL 103
PP++ ++ PDV+ D KDEFLV+ CDGIW++ ++ +++ F++ + L I+
Sbjct 280 PPQEAQVTVEPDVLMHKIDYSKDEFLVLACDGIWDIYNNKQLIHFIKYHLVSGTKLDTII 339
Query 104 KELFDSLISPNPALFEYGCDNMTAFIVDLQ 133
+L D I+ + G DNMTA IV L
Sbjct 340 TKLLDHGIAQANSNTGVGFDNMTAIIVVLN 369
> Hs17466020
Length=228
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 34/106 (32%), Positives = 54/106 (50%), Gaps = 4/106 (3%)
Query 15 RISRNGSCKWELELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIGCD 74
I+ N + L + K G YK P Q++S P+V R E D F+++ CD
Sbjct 43 HIADNQDLQASLAVSKALGDFDYKCIHGKGPTGQLVSPEPEVHDTERSEEDDHFIILACD 102
Query 75 GIWELLSSAEVVEFVRRRIEHTPDLCQILKELFDSL----ISPNPA 116
IW+++ + E+ +FVR R+E T D ++ E+ D+ ISP A
Sbjct 103 SIWDVMGNEELCDFVRSRLEVTDDPEKVYNEIVDTCLHKGISPEAA 148
> Hs4505995
Length=479
Score = 62.4 bits (150), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 37/112 (33%), Positives = 63/112 (56%), Gaps = 11/112 (9%)
Query 19 NGSCKWELELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIGCDGIWE 78
NGS L + + G YK D P +Q++S P+V + R E+DEF+++ CDGIW+
Sbjct 193 NGS----LAVSRALGDYDYKCVDGKGPTEQLVSPEPEVYEILR-AEEDEFIILACDGIWD 247
Query 79 LLSSAEVVEFVRRRIEHTPDLCQILKELFDSLISPNPALFEYGCDNMTAFIV 130
++S+ E+ E+V+ R+E + DL + + D+ L + DNM+ +V
Sbjct 248 VMSNEELCEYVKSRLEVSDDLENVCNWVVDT------CLHKGSRDNMSIVLV 293
> At2g25620
Length=392
Score = 59.3 bits (142), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 32/80 (40%), Positives = 47/80 (58%), Gaps = 7/80 (8%)
Query 54 PDVVTVNRDPEKDEFLVIGCDGIWELLSSAEVVEFVRRRIEHTPDLCQILKELFDSLISP 113
P+++T + E+DEFL+IGCDG+W++ S V+F RRR++ D KEL +
Sbjct 285 PELMTT-KLTEEDEFLIIGCDGVWDVFMSQNAVDFARRRLQEHNDPVMCSKELVEE---- 339
Query 114 NPALFEYGCDNMTAFIVDLQ 133
AL DN+TA +V LQ
Sbjct 340 --ALKRKSADNVTAVVVCLQ 357
> At3g62260
Length=383
Score = 55.8 bits (133), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 43/133 (32%), Positives = 61/133 (45%), Gaps = 18/133 (13%)
Query 13 GGRISRNGSCKWELELVKGFG----RLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEF 68
GG I+ +G L + + G +L + + L E +I +T+ D DEF
Sbjct 239 GGFITNDGYLNEVLAVTRALGDWDLKLPHGSQSPLISEPEIKQ-----ITLTED---DEF 290
Query 69 LVIGCDGIWELLSSAEVVEFVRRRIEHTPDLCQILKELFDSLISPNPALFEYGCDNMTAF 128
LVIGCDGIW++L+S E V VRR + D + +EL AL DN+TA
Sbjct 291 LVIGCDGIWDVLTSQEAVSIVRRGLNRHNDPTRCARELV------MEALGRNSFDNLTAV 344
Query 129 IVDLQAEKRRQLP 141
+V R P
Sbjct 345 VVCFMTMDRGDKP 357
> At1g18030
Length=341
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 34/119 (28%), Positives = 59/119 (49%), Gaps = 16/119 (13%)
Query 13 GGRISRNGSCKWELELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIG 72
GG IS NG + LE+ + FG +K K +S PD+ E++ F+++G
Sbjct 233 GGVISSNGRLQGRLEVSRAFGDRHFK--------KFGVSATPDIHAFEL-TERENFMILG 283
Query 73 CDGIWELLSSAEVVEFVRRRIEHTPDLCQILKELFDSLISPNPALFEYGC-DNMTAFIV 130
CDG+WE+ ++ V FV++ ++ + + + L A+ E C DN TA ++
Sbjct 284 CDGLWEVFGPSDAVGFVQKLLKEGLHVSTVSRRLV------KEAVKERRCKDNCTAIVI 336
> 7301795
Length=368
Score = 52.4 bits (124), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 29/105 (27%), Positives = 54/105 (51%), Gaps = 7/105 (6%)
Query 26 LELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEV 85
L + + G +K +Q++S P++ +R + DEFLV+ CDGIW+++S+ +V
Sbjct 185 LAVSRALGDYDFKNVKEKGQCEQLVSPEPEIFCQSRQ-DSDEFLVLACDGIWDVMSNEDV 243
Query 86 VEFVRRRIEHTPDLCQILKELFDSLISPNPALFEYGCDNMTAFIV 130
F+ R+ T +L I ++ D+ + DNM+ I+
Sbjct 244 CSFIHSRMRVTSNLVSIANQVVDTCLHKGSR------DNMSIIII 282
> At3g51470
Length=361
Score = 48.5 bits (114), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 26/71 (36%), Positives = 41/71 (57%), Gaps = 14/71 (19%)
Query 64 EKDEFLVIGCDGIWELLSSAEVVEFVRRRI--EHTPDLCQ--ILKELFDSLISPNPALFE 119
E+DE+L++GCDG+W+++SS V VRR + + P+ C ++KE AL
Sbjct 265 EEDEYLIMGCDGLWDVMSSQCAVTMVRRELMQHNDPERCSQALVKE----------ALQR 314
Query 120 YGCDNMTAFIV 130
CDN+T +V
Sbjct 315 NSCDNLTVVVV 325
> Hs21245120
Length=181
Score = 47.4 bits (111), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 34/132 (25%), Positives = 62/132 (46%), Gaps = 24/132 (18%)
Query 13 GGRISRNGSCKWE--LELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLV 70
GG IS NGS + + L + + G K + + P+ PD++T + D + EF++
Sbjct 67 GGFISFNGSWRVQGILAMSRSLGDYPLKNLNVVIPD-------PDILTFDLDKLQPEFMI 119
Query 71 IGCDGIWELLSSAEVVEFVRRRIE--HTPDLCQILKELFDSLISPNPALFEYGC-DNMTA 127
+ DG+W+ S+ E V F++ R++ H +L+ F GC DN+T
Sbjct 120 LASDGLWDAFSNEEAVRFIKERLDEPHFGAKSIVLQS------------FYRGCPDNITV 167
Query 128 FIVDLQAEKRRQ 139
+V + + +
Sbjct 168 MVVKFRNSSKTE 179
> At2g33700
Length=380
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 38/69 (55%), Gaps = 10/69 (14%)
Query 64 EKDEFLVIGCDGIWELLSSAEVVEFVRRR--IEHTPDLCQILKELFDSLISPNPALFEYG 121
E DEFL++GCDG+W+++SS V R+ I + P+ C +EL + N
Sbjct 282 EDDEFLIMGCDGLWDVMSSQCAVTIARKELMIHNDPERCS--RELVREALKRNT------ 333
Query 122 CDNMTAFIV 130
CDN+T +V
Sbjct 334 CDNLTVIVV 342
> Hs18589868
Length=263
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 38/71 (53%), Gaps = 2/71 (2%)
Query 13 GGRISRNGSCKWELELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIG 72
GG I R + L + + G YK PPE Q++S P+V + R E DEF+++
Sbjct 137 GGTIRRR-RVEGSLAVSRALGDFTYKEAPGRPPELQLVSAEPEVAALARQAE-DEFMLLA 194
Query 73 CDGIWELLSSA 83
DG+W+ +S A
Sbjct 195 SDGVWDTVSGA 205
> At3g17250
Length=378
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 16/35 (45%), Positives = 26/35 (74%), Gaps = 0/35 (0%)
Query 64 EKDEFLVIGCDGIWELLSSAEVVEFVRRRIEHTPD 98
E+DEFL++GCDG+W++++S V FVR+ + D
Sbjct 287 EEDEFLIMGCDGVWDVMTSQYAVTFVRQGLRRHGD 321
> SPCC4F11.02
Length=347
Score = 42.7 bits (99), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 41/86 (47%), Gaps = 10/86 (11%)
Query 47 KQILSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEVVEFVRRRIEHTPDLCQILKEL 106
K+++S P DEF +I CDG+W+++S E V+FVR + +P +
Sbjct 248 KELVSAHPFTTETRIWNGHDEFFIIACDGLWDVVSDQEAVDFVRNFV--SPREAAVRLVE 305
Query 107 FDSLISPNPALFEYGCDNMTAFIVDL 132
F AL DN+T +V+L
Sbjct 306 F--------ALKRLSTDNITCIVVNL 323
> At5g02760
Length=361
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 48/97 (49%), Gaps = 11/97 (11%)
Query 47 KQILSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEVVEFV---------RRRIEHTP 97
K ILS P V T+ R +DEF+++ DG+WE LS+ E V+ V RR ++
Sbjct 241 KPILSADPSV-TITRLSPQDEFIILASDGLWEHLSNQEAVDIVHNSPRQGIARRLLKAAL 299
Query 98 DLCQILKEL-FDSLISPNPALFEYGCDNMTAFIVDLQ 133
+E+ + L +P + + D++T +V L
Sbjct 300 KEAAKKREMRYSDLTEIHPGVRRHFHDDITVIVVYLN 336
> At1g48040
Length=377
Score = 42.4 bits (98), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 25/67 (37%), Positives = 33/67 (49%), Gaps = 6/67 (8%)
Query 64 EKDEFLVIGCDGIWELLSSAEVVEFVRRRIEHTPDLCQILKELFDSLISPNPALFEYGCD 123
E DEFL++ CDGIW++LSS V VR+ + D Q EL A D
Sbjct 281 EDDEFLILACDGIWDVLSSQNAVSNVRQGLRRHGDPRQCAMEL------GKEAARLQSSD 334
Query 124 NMTAFIV 130
NMT ++
Sbjct 335 NMTVIVI 341
> At1g17550
Length=527
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 36/140 (25%), Positives = 61/140 (43%), Gaps = 25/140 (17%)
Query 14 GRISRNGSCKWELELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIGC 73
G ++ + CK GF + D L P + P+V + R E DE L++
Sbjct 392 GVLAMSSLCKIMRSPESGFQSASFSGDQYLEP---FVIPDPEVTFMPRARE-DECLILAS 447
Query 74 DGIWELLSSAEVVEFVRRRIE--HTPD--------------LCQILKELFDSLISPNPAL 117
DG+W+++S+ E +F RRRI H + CQ E L A+
Sbjct 448 DGLWDVMSNQEACDFARRRILAWHKKNGALPLAERGVGEDQACQAAAEYLSKL-----AI 502
Query 118 FEYGCDNMTAFIVDLQAEKR 137
DN++ ++DL+A+++
Sbjct 503 QMGSKDNISIIVIDLKAQRK 522
> SPAC10F6.17c
Length=350
Score = 41.6 bits (96), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 17/67 (25%), Positives = 35/67 (52%), Gaps = 0/67 (0%)
Query 23 KWELELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSS 82
KW E+ + R + A ++ P++ ++ +P+K FL++ DG+W+ +SS
Sbjct 199 KWSQEISERLHREYFSASPIPVKTPPYVTAVPEIESITVNPKKHRFLIMASDGLWDTMSS 258
Query 83 AEVVEFV 89
+ V+ V
Sbjct 259 EQAVQLV 265
> At4g27800
Length=388
Score = 40.8 bits (94), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 28/113 (24%), Positives = 54/113 (47%), Gaps = 13/113 (11%)
Query 33 GRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEVVEFVRRR 92
GR K + + ++ PD+ V + EF+++ DG+W+ + S++VV +VR +
Sbjct 256 GRWSEKFVSRIEFKGDMVVATPDIFQVPLTSDV-EFIILASDGLWDYMKSSDVVSYVRDQ 314
Query 93 IEHTPDL---CQILKELFDSLISPNPALFEYGCDNMTAFIVDLQAEKRRQLPA 142
+ ++ C+ L ++ AL DN++ I DL + + LPA
Sbjct 315 LRKHGNVQLACESLAQV---------ALDRRSQDNISIIIADLGRTEWKNLPA 358
> 7290827
Length=475
Score = 40.0 bits (92), Expect = 0.001, Method: Composition-based stats.
Identities = 30/105 (28%), Positives = 42/105 (40%), Gaps = 18/105 (17%)
Query 6 GEDPDIRGGRISRNGSCKWELELVKGFGRLGYKADDSLPPEKQI---------------- 49
E P + RNG +L ++ FG YK + +K +
Sbjct 263 AEHPKEEHETVIRNGRLLSQLAPLRAFGDFRYKWSQEIMQQKVLPMFGVQAMAPNYYTPP 322
Query 50 -LSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEVVEFVRRRI 93
L+ PDV P D+FLVI DG+W+ L +EVV V I
Sbjct 323 YLTARPDVQQHELGP-NDKFLVIASDGLWDFLPPSEVVSLVGEHI 366
> At5g36250
Length=448
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/42 (47%), Positives = 28/42 (66%), Gaps = 1/42 (2%)
Query 50 LSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEVVEFVRR 91
L PDV + R EKDEF+V+ DGIW+ L++ EVV+ V +
Sbjct 306 LISVPDV-SYRRLTEKDEFVVLATDGIWDALTNEEVVKIVAK 346
> At1g72770
Length=511
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 29/100 (29%), Positives = 49/100 (49%), Gaps = 22/100 (22%)
Query 54 PDVVTVNRDPEKDEFLVIGCDGIWELLSSAEVVEFVRRRI------EHTPDL-------- 99
P+V + R E DE L++ DG+W+++++ EV E RRRI P L
Sbjct 413 PEVTFMPRSRE-DECLILASDGLWDVMNNQEVCEIARRRILMWHKKNGAPPLAERGKGID 471
Query 100 --CQILKELFDSLISPNPALFEYGCDNMTAFIVDLQAEKR 137
CQ + L AL + DN++ ++DL+A+++
Sbjct 472 PACQAAADYLSML-----ALQKGSKDNISIIVIDLKAQRK 506
> At5g66080
Length=385
Score = 39.7 bits (91), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 19/49 (38%), Positives = 29/49 (59%), Gaps = 1/49 (2%)
Query 44 PPEKQILSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEVVEFVRRR 92
P ++ ILS P + + P+ D+FL+ DG+WE LS+ E VE V+
Sbjct 259 PMKRPILSWEPSITVHDLQPD-DQFLIFASDGLWEQLSNQEAVEIVQNH 306
> Hs16164686
Length=529
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 28/91 (30%), Positives = 43/91 (47%), Gaps = 13/91 (14%)
Query 23 KWELELVKGFGRLGYKAD-----DSLPPE---KQILSGCPDVVTVNRDPEKDEFLVIGCD 74
KW EL + G+ + PP L+ P+V T +R +D+FLV+ D
Sbjct 354 KWSKELQRSILERGFNTEALNIYQFTPPHYYTPPYLTAEPEV-TYHRLRPQDKFLVLASD 412
Query 75 GIWELLSSAEVVEFVRRRIE----HTPDLCQ 101
G+W++LS+ +VV V + H DL Q
Sbjct 413 GLWDMLSNEDVVRLVVGHLAEADWHKTDLAQ 443
> At3g51370
Length=379
Score = 38.5 bits (88), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 29/49 (59%), Gaps = 1/49 (2%)
Query 44 PPEKQILSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEVVEFVRRR 92
P ++ ILSG P + P+ D+FL+ DG+WE +S+ E V+ V+
Sbjct 256 PFKRPILSGEPTITEHEIQPQ-DKFLIFASDGLWEQMSNQEAVDIVQNH 303
> At3g16800
Length=351
Score = 38.5 bits (88), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 25/34 (73%), Gaps = 0/34 (0%)
Query 57 VTVNRDPEKDEFLVIGCDGIWELLSSAEVVEFVR 90
VT + +KD+FL++ DG+W+++++ E VE VR
Sbjct 276 VTYRKITDKDQFLILATDGMWDVMTNNEAVEIVR 309
> Hs20558275
Length=1205
Score = 38.5 bits (88), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 29/80 (36%), Positives = 41/80 (51%), Gaps = 13/80 (16%)
Query 54 PDVVTVNRDPEKDEFLVIGCDGIWELLSSAEVVEFVRRRIEHTPDLCQILKELFDSLISP 113
P V +V P+ DEF ++G G+W+ LS E VE VR + PD K+L S
Sbjct 843 PHVQSVLLTPQ-DEFFILGSKGLWDSLSVEEAVEAVR----NVPDALAAAKKLCTLAQS- 896
Query 114 NPALFEYGC-DNMTAFIVDL 132
YGC D+++A +V L
Sbjct 897 ------YGCHDSISAVVVQL 910
> At5g26010
Length=337
Score = 37.7 bits (86), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 24/33 (72%), Gaps = 0/33 (0%)
Query 57 VTVNRDPEKDEFLVIGCDGIWELLSSAEVVEFV 89
++ +R KD+FLV+ DG+W++LS+ EVV +
Sbjct 257 ISQHRITSKDQFLVLATDGVWDMLSNDEVVSLI 289
> At3g02750
Length=492
Score = 37.7 bits (86), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 19/40 (47%), Positives = 27/40 (67%), Gaps = 1/40 (2%)
Query 50 LSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEVVEFV 89
L PDV + + EKDEF+V+ DGIW++LS+ +VV V
Sbjct 315 LISVPDV-SFRQLTEKDEFIVLATDGIWDVLSNEDVVAIV 353
> At3g55050
Length=409
Score = 37.4 bits (85), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 28/44 (63%), Gaps = 1/44 (2%)
Query 46 EKQILSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEVVEFV 89
EK I+ P +TV++ +D+FL+ DG+WE LS+ E V+ V
Sbjct 288 EKPIMRAEP-TITVHKIHPEDQFLIFASDGLWEHLSNQEAVDIV 330
> 7290521
Length=1400
Score = 37.4 bits (85), Expect = 0.010, Method: Composition-based stats.
Identities = 29/103 (28%), Positives = 48/103 (46%), Gaps = 16/103 (15%)
Query 49 ILSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEVVEFVRRRIEHTPDLCQILKELFD 108
++S PDV V +P L+ G DG+W ++++ E V+ VR+ EH + +IL E
Sbjct 453 VVSPDPDVKVVKINPSTFRCLIFGTDGLWNVVTAQEAVDSVRK--EHL--IGEILNE--Q 506
Query 109 SLISPNPALFEYG----------CDNMTAFIVDLQAEKRRQLP 141
+++P+ AL + DN + V L R P
Sbjct 507 DVMNPSKALVDQALKTWAAKKMRADNTSVVTVILTPAARNNSP 549
> 7297303
Length=524
Score = 37.0 bits (84), Expect = 0.012, Method: Composition-based stats.
Identities = 15/54 (27%), Positives = 28/54 (51%), Gaps = 1/54 (1%)
Query 46 EKQILSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEVVEFVRRRIEHTPDL 99
+K ++ PD++T + K FL++ DG+W+ S+ E F ++ PD
Sbjct 405 DKNLVIATPDILTFELNDHKPHFLILASDGLWDTFSNEEACTFALEHLKE-PDF 457
> At3g12620
Length=376
Score = 37.0 bits (84), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 27/44 (61%), Gaps = 1/44 (2%)
Query 46 EKQILSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEVVEFV 89
K IL P +TV++ +D+FL+ DG+WE LS+ E V+ V
Sbjct 253 HKPILRAEP-AITVHKIHPEDQFLIFASDGLWEHLSNQEAVDIV 295
> Hs17455719
Length=514
Score = 36.6 bits (83), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 33/65 (50%), Gaps = 3/65 (4%)
Query 28 LVKGFGRLGYKADDSLPPEKQILSGCPDVVTVN---RDPEKDEFLVIGCDGIWELLSSAE 84
+ +G G K DS K LS P+V + D D+ L++ DG+W++LS+ E
Sbjct 388 VTRGLGDHDLKVHDSNIYIKPFLSSAPEVRIYDLSKYDHGSDDVLILATDGLWDVLSNEE 447
Query 85 VVEFV 89
V E +
Sbjct 448 VAEAI 452
> At3g63340_1
Length=750
Score = 36.2 bits (82), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 34/122 (27%), Positives = 51/122 (41%), Gaps = 18/122 (14%)
Query 25 ELELVKGFGRLGYKADDSLPPEKQILSGCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAE 84
+L + + G L Y++ + P+V+ D FLV+ DGI+E L E
Sbjct 318 QLTVSRAIGDLTYRSYGVI--------SAPEVMDWQPLVANDSFLVVSSDGIFEKLEVQE 369
Query 85 VVEFVRRRIEHT------PDLCQILKELFDSLISPNPALFEYGCDNMTAFIVDLQAEKRR 138
V + + T P C I L D L+ N A + DNM A +V L++
Sbjct 370 VCDLLWEVNNQTSSGAGVPSYCSI--SLADCLV--NTAFEKGSMDNMAAVVVPLKSNLVT 425
Query 139 QL 140
QL
Sbjct 426 QL 427
> At4g11040
Length=295
Score = 36.2 bits (82), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 19/86 (22%), Positives = 45/86 (52%), Gaps = 3/86 (3%)
Query 52 GCPDVVTVNRDPEKDEFLVIGCDGIWELLSSAEVVEFVRRRIEHT--PDLC-QILKELFD 108
G ++ +++ + D+F+V+ CDG+W+++S + + V+R + PD C
Sbjct 206 GVEQILKIHKRKKIDDFIVLACDGLWDVVSDDDTYQLVKRCLYGKLPPDGCISESSSTKA 265
Query 109 SLISPNPALFEYGCDNMTAFIVDLQA 134
++I A+ +N+ ++DL++
Sbjct 266 AVILAELAIARGSKENINVIVIDLKS 291
Lambda K H
0.319 0.140 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1675978996
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40