bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1488_orf2
Length=94
Score E
Sequences producing significant alignments: (Bits) Value
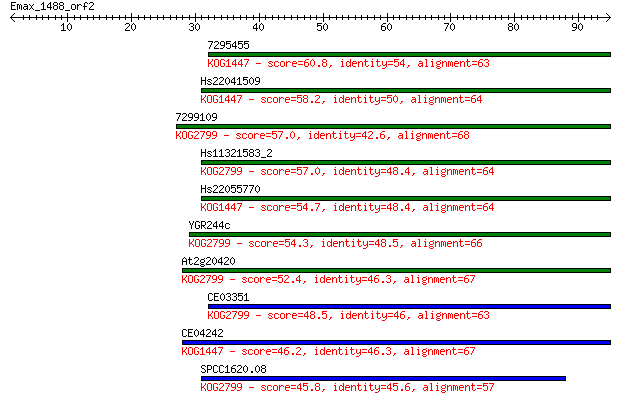
7295455 60.8 5e-10
Hs22041509 58.2 4e-09
7299109 57.0 7e-09
Hs11321583_2 57.0 8e-09
Hs22055770 54.7 4e-08
YGR244c 54.3 5e-08
At2g20420 52.4 2e-07
CE03351 48.5 3e-06
CE04242 46.2 1e-05
SPCC1620.08 45.8 2e-05
> 7295455
Length=416
Score = 60.8 bits (146), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 34/63 (53%), Positives = 41/63 (65%), Gaps = 1/63 (1%)
Query 32 VVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGRKVRR 91
VVKAQILAGGR G F +NG+KGGVH N V S+ MI LITKQT K G V +
Sbjct 66 VVKAQILAGGRGKGTF-DNGFKGGVHITTNKSEVLSLTQQMIGNRLITKQTPKSGILVNK 124
Query 92 LLI 94
+++
Sbjct 125 VMV 127
> Hs22041509
Length=432
Score = 58.2 bits (139), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 32/64 (50%), Positives = 43/64 (67%), Gaps = 1/64 (1%)
Query 31 LVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGRKVR 90
+V+KAQILAGGR G F +G KGGVH +P +V +A MI L TKQT K+G KV
Sbjct 80 IVLKAQILAGGRGKGVFN-SGLKGGVHLTKDPNVVGQLAKQMIGYNLATKQTPKEGVKVN 138
Query 91 RLLI 94
++++
Sbjct 139 KVMV 142
> 7299109
Length=465
Score = 57.0 bits (136), Expect = 7e-09, Method: Composition-based stats.
Identities = 29/68 (42%), Positives = 46/68 (67%), Gaps = 1/68 (1%)
Query 27 QENALVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKG 86
+ + LV+KAQ+LAGGR G FK NG KGGV +P+ +++ MI++ L+TKQT G
Sbjct 76 KTDNLVLKAQVLAGGRGKGTFK-NGLKGGVRVVYDPQTAEELSSKMIDQLLVTKQTGAAG 134
Query 87 RKVRRLLI 94
R +++++
Sbjct 135 RICKKVMV 142
> Hs11321583_2
Length=385
Score = 57.0 bits (136), Expect = 8e-09, Method: Composition-based stats.
Identities = 31/64 (48%), Positives = 44/64 (68%), Gaps = 1/64 (1%)
Query 31 LVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGRKVR 90
+V+KAQ+LAGGR G F E+G KGGV +P+ +V++ MI K L TKQT +KGR
Sbjct 17 VVIKAQVLAGGRGKGTF-ESGLKGGVKIVFSPEEAKAVSSQMIGKKLFTKQTGEKGRICN 75
Query 91 RLLI 94
++L+
Sbjct 76 QVLV 79
> Hs22055770
Length=383
Score = 54.7 bits (130), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 31/64 (48%), Positives = 42/64 (65%), Gaps = 1/64 (1%)
Query 31 LVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGRKVR 90
+V+KAQILAGGR G F +G K GVH +P +V +A MI L TKQT K+G KV
Sbjct 32 IVLKAQILAGGRGKGVFN-SGLKRGVHLTKDPNVVGQLAKQMIGYNLATKQTPKEGVKVN 90
Query 91 RLLI 94
++++
Sbjct 91 KVMV 94
> YGR244c
Length=427
Score = 54.3 bits (129), Expect = 5e-08, Method: Composition-based stats.
Identities = 32/66 (48%), Positives = 39/66 (59%), Gaps = 1/66 (1%)
Query 29 NALVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGRK 88
N LV+KAQ L GGR G F + GYK GVH +P+ VA M+N LITKQT G+
Sbjct 71 NKLVIKAQALTGGRGKGHF-DTGYKSGVHMIESPQQAEDVAKEMLNHNLITKQTGIAGKP 129
Query 89 VRRLLI 94
V + I
Sbjct 130 VSAVYI 135
> At2g20420
Length=421
Score = 52.4 bits (124), Expect = 2e-07, Method: Composition-based stats.
Identities = 31/67 (46%), Positives = 44/67 (65%), Gaps = 2/67 (2%)
Query 28 ENALVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGR 87
E+ LVVK+QILAGGR LG FK +G KGGVH + +A M+ + L+TKQT +G+
Sbjct 68 ESELVVKSQILAGGRGLGTFK-SGLKGGVH-IVKRDEAEEIAGKMLGQVLVTKQTGPQGK 125
Query 88 KVRRLLI 94
V ++ +
Sbjct 126 VVSKVYL 132
> CE03351
Length=435
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 29/63 (46%), Positives = 37/63 (58%), Gaps = 1/63 (1%)
Query 32 VVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGRKVRR 91
VVKAQ+LAGGR G F +G +GGV P V A MI LITKQT +G+K
Sbjct 67 VVKAQVLAGGRGKGRFS-SGLQGGVQIVFTPDEVKQKAGMMIGANLITKQTDHRGKKCEE 125
Query 92 LLI 94
+++
Sbjct 126 VMV 128
> CE04242
Length=415
Score = 46.2 bits (108), Expect = 1e-05, Method: Composition-based stats.
Identities = 31/69 (44%), Positives = 40/69 (57%), Gaps = 3/69 (4%)
Query 28 ENALVVKAQILAGGRSLGFFKENGYKG--GVHACINPKLVFSVANNMINKTLITKQTTKK 85
++ VVKAQILAGGR G F NG KG GV + MI K L+TKQTT +
Sbjct 59 DHEYVVKAQILAGGRGKGKFI-NGTKGIGGVFITKEKDAALEAIDEMIGKRLVTKQTTSE 117
Query 86 GRKVRRLLI 94
G +V +++I
Sbjct 118 GVRVDKVMI 126
> SPCC1620.08
Length=433
Score = 45.8 bits (107), Expect = 2e-05, Method: Composition-based stats.
Identities = 26/57 (45%), Positives = 33/57 (57%), Gaps = 1/57 (1%)
Query 31 LVVKAQILAGGRSLGFFKENGYKGGVHACINPKLVFSVANNMINKTLITKQTTKKGR 87
LVVKAQ+LAGGR G F ++G +GGV + A MI LIT+QT G+
Sbjct 65 LVVKAQVLAGGRGKGQF-DSGLRGGVRPVYDATEARMFAEQMIGHKLITRQTGPAGK 120
Lambda K H
0.316 0.129 0.350
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164659894
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40