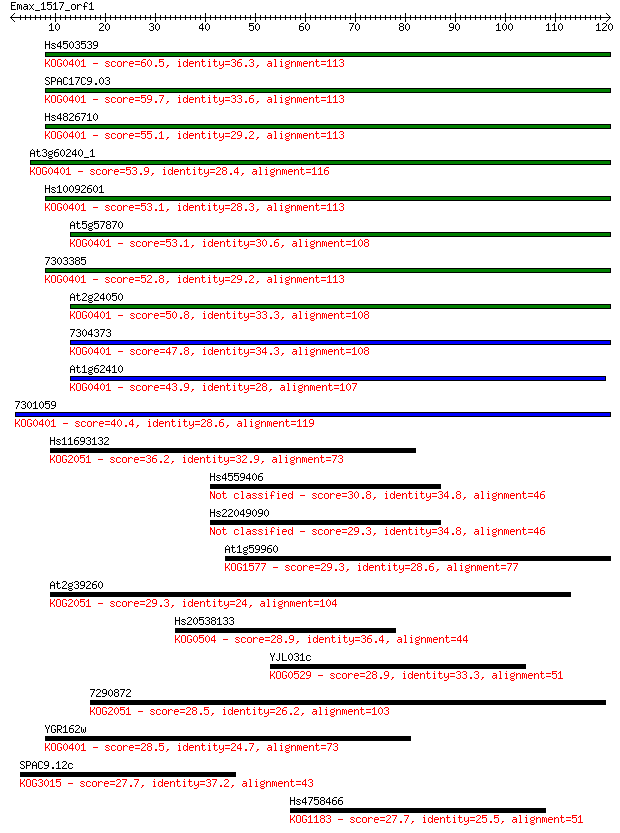
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1517_orf1
Length=120
Score E
Sequences producing significant alignments: (Bits) Value
Hs4503539 60.5 8e-10
SPAC17C9.03 59.7 1e-09
Hs4826710 55.1 3e-08
At3g60240_1 53.9 7e-08
Hs10092601 53.1 1e-07
At5g57870 53.1 1e-07
7303385 52.8 2e-07
At2g24050 50.8 7e-07
7304373 47.8 5e-06
At1g62410 43.9 7e-05
7301059 40.4 9e-04
Hs11693132 36.2 0.015
Hs4559406 30.8 0.67
Hs22049090 29.3 1.7
At1g59960 29.3 1.8
At2g39260 29.3 1.9
Hs20538133 28.9 2.3
YJL031c 28.9 2.7
7290872 28.5 2.7
YGR162w 28.5 3.3
SPAC9.12c 27.7 5.0
Hs4758466 27.7 5.9
> Hs4503539
Length=907
Score = 60.5 bits (145), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 41/114 (35%), Positives = 60/114 (52%), Gaps = 9/114 (7%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
K M GN+ F+GEL K +I ++IL +CI LL+K+ KD G +E LC +
Sbjct 204 KIKMLGNIKFIGELGKLDLIHESILHKCIKTLLEKKKRV----QLKDMG-EDLECLCQIM 258
Query 68 HTVGPFFDNPQWR-LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
TVGP D+ + + L ++ RM SL LP R+R L+ D ++ R W
Sbjct 259 RTVGPRLDHERAKSLMDQYFARM---CSLMLSKELPARIRFLLQDTVELREHHW 309
> SPAC17C9.03
Length=1403
Score = 59.7 bits (143), Expect = 1e-09, Method: Composition-based stats.
Identities = 38/115 (33%), Positives = 57/115 (49%), Gaps = 14/115 (12%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
K+ G + F+GEL K M+S+ I+ ECI RLL N D +E+LC L
Sbjct 1144 KRRGLGLVRFIGELFKLSMLSEKIMHECIKRLL---------GNVTDPEEEEIESLCRLL 1194
Query 68 HTVGPFFDNPQ--WRLYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
TVG D + + RM+ + + +LP R++ ++ DV+DSR GW
Sbjct 1195 MTVGVNIDATEKGHAAMDVYVLRMETITKI---PNLPSRIKFMLMDVMDSRKNGW 1246
> Hs4826710
Length=1396
Score = 55.1 bits (131), Expect = 3e-08, Method: Composition-based stats.
Identities = 33/114 (28%), Positives = 58/114 (50%), Gaps = 16/114 (14%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
++ GN+ F+GEL K KM+++AI+ +C+ +LL+ E +E LC L
Sbjct 696 RRCSLGNIKFIGELFKLKMLTEAIMHDCVVKLLKNHDE------------ESLECLCRLL 743
Query 68 HTVGPFFDNPQWR-LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
T+G D + + ++ +M+ + SS R+R ++ DVLD R + W
Sbjct 744 TTIGKDLDFEKAKPRMDQYFNQMEKIIKEKKTSS---RIRFMLQDVLDLRGSNW 794
> At3g60240_1
Length=1528
Score = 53.9 bits (128), Expect = 7e-08, Method: Composition-based stats.
Identities = 33/118 (27%), Positives = 61/118 (51%), Gaps = 15/118 (12%)
Query 5 LNYKKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALC 64
L ++ M GN+ +GEL K +M+++ I+ CI +LL ++D ++EALC
Sbjct 1033 LQVRRRMLGNIRLIGELYKKRMLTEKIMHACIQKLL---------GYNQDPHEENIEALC 1083
Query 65 AFLHTVGPFFDN--PQWRLYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
+ T+G D+ ++++ G F +M + L+ L RVR ++ + +D R W
Sbjct 1084 KLMSTIGVMIDHNKAKFQMDGYF-EKMKM---LSCKQELSSRVRFMLINAIDLRKNKW 1137
> Hs10092601
Length=1585
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 32/114 (28%), Positives = 57/114 (50%), Gaps = 16/114 (14%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
++ GN+ F+GEL K KM+++AI+ +C+ +LL+ E +E LC L
Sbjct 886 RRRSIGNIKFIGELFKLKMLTEAIMHDCVVKLLKNHDE------------ESLECLCRLL 933
Query 68 HTVGPFFDNPQWR-LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
T+G D + + ++ +M+ + SS R+R ++ DV+D R W
Sbjct 934 TTIGKDLDFEKAKPRMDQYFNQMEKIVKERKTSS---RIRFMLQDVIDLRLCNW 984
> At5g57870
Length=780
Score = 53.1 bits (126), Expect = 1e-07, Method: Composition-based stats.
Identities = 33/111 (29%), Positives = 56/111 (50%), Gaps = 14/111 (12%)
Query 13 GNMIFVGELLKSKMISQAILLECIDRLLQKRAE-CIAASNSKDQGIHHVEALCAFLHTVG 71
GN+ +GELLK KM+ + I+ + LL + C A N VEA+C F T+G
Sbjct 343 GNIRLIGELLKQKMVPEKIVHHIVQELLGADEKVCPAEEN--------VEAICHFFKTIG 394
Query 72 PFFDN--PQWRLYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
D R+ + +R+ +L+ + L R+R ++ +++D R+ GW
Sbjct 395 KQLDGNVKSKRINDVYFKRLQ---ALSKNPQLELRLRFMVQNIIDMRSNGW 442
> 7303385
Length=869
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 33/114 (28%), Positives = 58/114 (50%), Gaps = 7/114 (6%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
K+ M GN+ F+GEL K M+S+ +L +CI L K+ + A + + +E L L
Sbjct 67 KQRMLGNVKFIGELNKLDMLSKNVLHQCIMELFDKKKKRTAGTQ---EMCEDMECLAQLL 123
Query 68 HTVGPFFDNPQWR-LYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
T G D+ Q + L ++ +++ + S P R+R ++ DV++ R W
Sbjct 124 KTCGKNLDSEQGKELMNQYFEKLE---RRSKSSEYPPRIRFMLKDVIELRQNNW 174
> At2g24050
Length=747
Score = 50.8 bits (120), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 36/111 (32%), Positives = 53/111 (47%), Gaps = 14/111 (12%)
Query 13 GNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFLHTVGP 72
GN+ +GELLK KM+ + I+ + LL + A VEALC F T+G
Sbjct 302 GNIRLIGELLKQKMVPEKIVHHIVQELLGDDTKACPAEGD-------VEALCQFFITIGK 354
Query 73 FFDN-PQWRLYGE--FCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAGW 120
D+ P+ R + F R + LA L R+R ++ +V+D RA W
Sbjct 355 QLDDSPRSRGINDTYFGR----LKELARHPQLELRLRFMVQNVVDLRANKW 401
> 7304373
Length=1666
Score = 47.8 bits (112), Expect = 5e-06, Method: Composition-based stats.
Identities = 37/119 (31%), Positives = 54/119 (45%), Gaps = 24/119 (20%)
Query 13 GNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFLHTVGP 72
G + F+GEL K M++ I+ CID LL +E + +E LC L TVG
Sbjct 895 GTVRFIGELFKISMLTGKIIYSCIDTLLNPHSEDM------------LECLCKLLTTVGA 942
Query 73 FFD-------NPQWRLYG--EFCRRMDLVASLASD--SSLPFRVRCLMSDVLDSRAAGW 120
F+ +P R Y + RM +AS + + RVR ++ DV+D R W
Sbjct 943 KFEKTPVNSKDPS-RCYSLEKSITRMQAIASKTDKDGARVSSRVRFMLQDVIDLRKNKW 1000
> At1g62410
Length=223
Score = 43.9 bits (102), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 30/107 (28%), Positives = 55/107 (51%), Gaps = 10/107 (9%)
Query 13 GNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFLHTVGP 72
GN+ F GEL +M+++ ++L +LL+ AE + S K + A+C FL+TVG
Sbjct 104 GNLRFCGELFLKRMLTEKVVLAIGQKLLED-AEQMCPSEEK------IIAICLFLNTVGK 156
Query 73 FFDNPQWRLYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAG 119
D+ +L E RR+ +L++ L +R ++ ++ + G
Sbjct 157 KLDSLNSKLMNEILRRL---KNLSNHPQLVMSLRLMVGKIIHLHSIG 200
> 7301059
Length=1905
Score = 40.4 bits (93), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 34/133 (25%), Positives = 57/133 (42%), Gaps = 26/133 (19%)
Query 2 EFELNYKKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVE 61
+ E +++ G + F+GEL K + ++ +L C++ LL+ E +E
Sbjct 1231 DLEYQFRRRAWGTVRFIGELFKLQSLTNDRVLNCVESLLEHGCE------------EKLE 1278
Query 62 ALCAFLHTVGPFFDN--PQW-----RLYGEFCRRMDLVASLASDS-------SLPFRVRC 107
+C L TVG ++ P+ R+ F R D++ S + RVR
Sbjct 1279 YMCKLLTTVGHLLESSLPEHYQLRDRIEKIFRRIQDIINRSRGTSHRQQVHIKISSRVRF 1338
Query 108 LMSDVLDSRAAGW 120
+M DVLD R W
Sbjct 1339 MMQDVLDLRLRNW 1351
> Hs11693132
Length=1272
Score = 36.2 bits (82), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 24/74 (32%), Positives = 33/74 (44%), Gaps = 13/74 (17%)
Query 9 KTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFLH 68
+T + F+GEL K KM ++ L C+ LL D HH+E C L
Sbjct 661 ETKNKTVRFIGELTKFKMFTKNDTLHCLKMLLS------------DFSHHHIEMACTLLE 708
Query 69 TVGPF-FDNPQWRL 81
T G F F +P+ L
Sbjct 709 TCGRFLFRSPESHL 722
> Hs4559406
Length=934
Score = 30.8 bits (68), Expect = 0.67, Method: Composition-based stats.
Identities = 16/46 (34%), Positives = 21/46 (45%), Gaps = 1/46 (2%)
Query 41 QKRAECIAASNSKDQGIHHVEALCAFLHTVGPFFDNPQWRLYGEFC 86
Q +ECI S +D HH E LC F +F +P + E C
Sbjct 779 QSGSECICMSPEEDCS-HHSEDLCVFDTDSNDYFTSPACKFLAEKC 823
> Hs22049090
Length=742
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 21/46 (45%), Gaps = 1/46 (2%)
Query 41 QKRAECIAASNSKDQGIHHVEALCAFLHTVGPFFDNPQWRLYGEFC 86
Q +ECI S +D HH E LC F +F +P + E C
Sbjct 587 QSGSECICMSPEEDCS-HHSEDLCVFDTDSNDYFTSPACKFLAEKC 631
> At1g59960
Length=326
Score = 29.3 bits (64), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 22/79 (27%), Positives = 32/79 (40%), Gaps = 16/79 (20%)
Query 44 AECIAASNSKDQGIHHVEALCAFLHTVGPFFDNPQW--RLYGEFCRRMDLVASLASDSSL 101
A+CI SN + + H+ ++ +V +P W R E CR D+V + S
Sbjct 163 AKCIGVSNFSCKKLQHILSIATIPPSVNQVEMSPIWQQRKLRELCRSNDIVVTAYS---- 218
Query 102 PFRVRCLMSDVLDSRAAGW 120
VL SR A W
Sbjct 219 ----------VLGSRGAFW 227
> At2g39260
Length=1029
Score = 29.3 bits (64), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 25/106 (23%), Positives = 46/106 (43%), Gaps = 17/106 (16%)
Query 9 KTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHH-VEALCAFL 67
+T N+ F+GEL K K++ ++ C+ L D+ HH ++ C L
Sbjct 402 ETKIRNIRFIGELCKFKIVPAGLVFSCLKACL-------------DEFTHHNIDVACNLL 448
Query 68 HTVGPF-FDNPQWRLYGEFCRRMDLVASLASDSSLPFRVRCLMSDV 112
T G F + +P+ L +D++ L + +L R L+ +
Sbjct 449 ETCGRFLYRSPETTL--RMTNMLDILMRLKNVKNLDPRQSTLVENA 492
> Hs20538133
Length=899
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 16/50 (32%), Positives = 22/50 (44%), Gaps = 6/50 (12%)
Query 34 ECIDRLLQKRAECIAASNSKDQGIH------HVEALCAFLHTVGPFFDNP 77
EC+D LLQ A+C+ + IH H+ L A L + NP
Sbjct 546 ECVDALLQHGAKCLLRDSRGRTPIHLSAACGHIGVLGALLQSAASMDANP 595
> YJL031c
Length=290
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 28/51 (54%), Gaps = 5/51 (9%)
Query 53 KDQGIHHVEALCAFLHTVGPFFDNPQWRLYGEFCRRMDLVASLASDSSLPF 103
+D+ I+ +EAL T NP++ + R D++ASLAS+ +PF
Sbjct 2 RDEKIYSIEALKK---TSELLEKNPEFNAIWNY--RRDIIASLASELEIPF 47
> 7290872
Length=1241
Score = 28.5 bits (62), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 27/103 (26%), Positives = 41/103 (39%), Gaps = 25/103 (24%)
Query 17 FVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFLHTVGPFFDN 76
F+GEL+K + + L C+ LL +D H +E CAF+ G + N
Sbjct 612 FIGELVKFGLFKKFDALGCLKMLL------------RDFQHHQIEMACAFVEVTGVYLYN 659
Query 77 PQWRLYGEFCRRMDLVASLASDSSLPFRVRCLMSDVLDSRAAG 119
CR L+ ++ D L R + +DSR A
Sbjct 660 ---------CRDARLLMNVFLDQML----RLKTATAMDSRHAA 689
> YGR162w
Length=952
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 18/73 (24%), Positives = 38/73 (52%), Gaps = 8/73 (10%)
Query 8 KKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAECIAASNSKDQGIHHVEALCAFL 67
K+ G + F+G L + +++ ++ EC RL++ + S S++ +E++ L
Sbjct 740 KRRGLGLVRFIGFLYRLNLLTGKMMFECFRRLMKD----LTDSPSEET----LESVVELL 791
Query 68 HTVGPFFDNPQWR 80
+TVG F+ +R
Sbjct 792 NTVGEQFETDSFR 804
> SPAC9.12c
Length=287
Score = 27.7 bits (60), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 23/43 (53%), Gaps = 0/43 (0%)
Query 3 FELNYKKTMTGNMIFVGELLKSKMISQAILLECIDRLLQKRAE 45
EL Y+ G++ E LK+K+ S AIL CI+ + K E
Sbjct 242 LELQYQTNRWGSLEDDNEDLKNKLASSAILSRCIEDMHDKSNE 284
> Hs4758466
Length=581
Score = 27.7 bits (60), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 13/53 (24%), Positives = 27/53 (50%), Gaps = 2/53 (3%)
Query 57 IHHVEALCAFLHTVGPFFDNPQWRLYGEFCRRMDLVASLASD--SSLPFRVRC 107
++H+ +++H + PF ++ W + C + + SL SD + L F + C
Sbjct 349 LYHIHLWISYIHLMSPFVEHILWHVGLSACLGLTVALSLLSDIIALLTFHIYC 401
Lambda K H
0.326 0.137 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160968786
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40