bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1535_orf1
Length=92
Score E
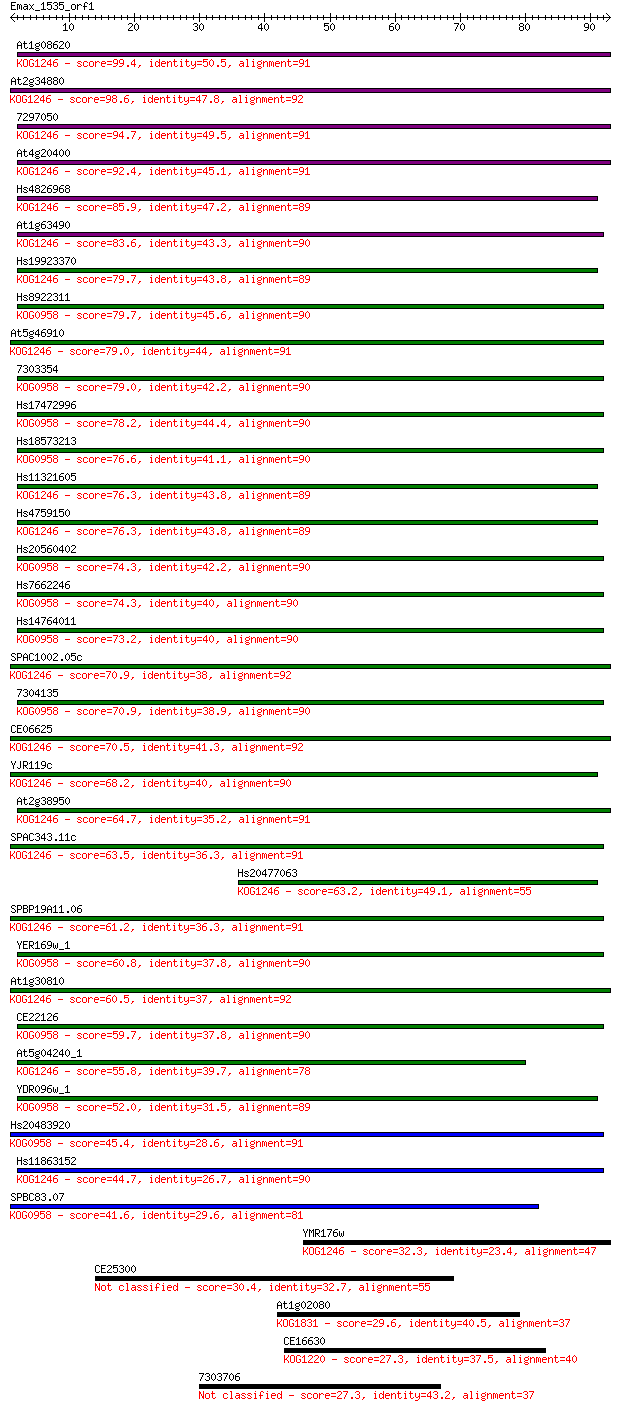
Sequences producing significant alignments: (Bits) Value
At1g08620 99.4 2e-21
At2g34880 98.6 2e-21
7297050 94.7 3e-20
At4g20400 92.4 2e-19
Hs4826968 85.9 2e-17
At1g63490 83.6 9e-17
Hs19923370 79.7 1e-15
Hs8922311 79.7 1e-15
At5g46910 79.0 2e-15
7303354 79.0 2e-15
Hs17472996 78.2 3e-15
Hs18573213 76.6 1e-14
Hs11321605 76.3 1e-14
Hs4759150 76.3 1e-14
Hs20560402 74.3 5e-14
Hs7662246 74.3 6e-14
Hs14764011 73.2 1e-13
SPAC1002.05c 70.9 5e-13
7304135 70.9 5e-13
CE06625 70.5 6e-13
YJR119c 68.2 3e-12
At2g38950 64.7 4e-11
SPAC343.11c 63.5 8e-11
Hs20477063 63.2 1e-10
SPBP19A11.06 61.2 5e-10
YER169w_1 60.8 6e-10
At1g30810 60.5 7e-10
CE22126 59.7 1e-09
At5g04240_1 55.8 2e-08
YDR096w_1 52.0 3e-07
Hs20483920 45.4 2e-05
Hs11863152 44.7 4e-05
SPBC83.07 41.6 3e-04
YMR176w 32.3 0.19
CE25300 30.4 0.75
At1g02080 29.6 1.2
CE16630 27.3 7.3
7303706 27.3 7.7
> At1g08620
Length=1183
Score = 99.4 bits (246), Expect = 2e-21, Method: Composition-based stats.
Identities = 46/93 (49%), Positives = 58/93 (62%), Gaps = 2/93 (2%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQSYLAD--RDPHYVLHSLTVQLPPSLFVENNIPVYRI 59
HWGAPK+WY V A +E ++ +L D + +LH L QL PS +PV+R
Sbjct 394 HWGAPKLWYGVGGKDAVKLEEAMRKHLPDLFEEQPDLLHKLVTQLSPSKLKTAGVPVHRC 453
Query 60 EQHAKEFVMLWPRTYHAGFNAGFNCNEACNFAP 92
QHA EFV+ +PR YHAGFN+GFNC EA N AP
Sbjct 454 VQHAGEFVLTFPRAYHAGFNSGFNCAEAVNVAP 486
> At2g34880
Length=806
Score = 98.6 bits (244), Expect = 2e-21, Method: Composition-based stats.
Identities = 44/95 (46%), Positives = 60/95 (63%), Gaps = 4/95 (4%)
Query 1 HHWGAPKIWYVVPPSRAPSVERTLQSYLAD---RDPHYVLHSLTVQLPPSLFVENNIPVY 57
HH+G PK+WY VP S A +E+ ++ +L D P +LH L Q P++ +PVY
Sbjct 319 HHFGEPKVWYGVPGSHATGLEKAMRKHLPDLFDEQPD-LLHELVTQFSPTILKNEGVPVY 377
Query 58 RIEQHAKEFVMLWPRTYHAGFNAGFNCNEACNFAP 92
R Q+A E+V+ +PR YH+GFN GFNC EA N AP
Sbjct 378 RAVQNAGEYVLTFPRAYHSGFNCGFNCAEAVNVAP 412
> 7297050
Length=1838
Score = 94.7 bits (234), Expect = 3e-20, Method: Composition-based stats.
Identities = 45/94 (47%), Positives = 58/94 (61%), Gaps = 4/94 (4%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQSY---LADRDPHYVLHSLTVQLPPSLFVENNIPVYR 58
HWG PK WY VP S A E T++ L P +LH L + P++ + N +PV+R
Sbjct 650 HWGEPKTWYGVPGSCAEQFEETMKQAAPELFSSQPD-LLHQLVTIMNPNILMNNRVPVFR 708
Query 59 IEQHAKEFVMLWPRTYHAGFNAGFNCNEACNFAP 92
+QHA EFV+ +PR YHAGFN G+N EA NFAP
Sbjct 709 TDQHAGEFVITFPRAYHAGFNQGYNFAEAVNFAP 742
> At4g20400
Length=872
Score = 92.4 bits (228), Expect = 2e-19, Method: Composition-based stats.
Identities = 41/93 (44%), Positives = 56/93 (60%), Gaps = 2/93 (2%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQSYLAD--RDPHYVLHSLTVQLPPSLFVENNIPVYRI 59
H G PK+WY +P + A S E ++ L D + +LH L QL P + E +PVYR
Sbjct 302 HTGDPKVWYGIPGNHAESFENVMKKRLPDLFEEQPDLLHQLVTQLSPRILKEEGVPVYRA 361
Query 60 EQHAKEFVMLWPRTYHAGFNAGFNCNEACNFAP 92
Q + EF++ +P+ YH+GFN GFNC EA N AP
Sbjct 362 VQRSGEFILTFPKAYHSGFNCGFNCAEAVNVAP 394
> Hs4826968
Length=1722
Score = 85.9 bits (211), Expect = 2e-17, Method: Composition-based stats.
Identities = 42/92 (45%), Positives = 55/92 (59%), Gaps = 4/92 (4%)
Query 2 HWGAPKIWYVVPPSRAPSVE---RTLQSYLADRDPHYVLHSLTVQLPPSLFVENNIPVYR 58
HWG PK WY VP A +E R L L + P +LH L + P++ +E+ +PVYR
Sbjct 496 HWGEPKTWYGVPSHAAEQLEEVMRELAPELFESQPD-LLHQLVTIMNPNVLMEHGVPVYR 554
Query 59 IEQHAKEFVMLWPRTYHAGFNAGFNCNEACNF 90
Q A EFV+ +PR YH+GFN G+N EA NF
Sbjct 555 TNQCAGEFVVTFPRAYHSGFNQGYNFAEAVNF 586
> At1g63490
Length=1324
Score = 83.6 bits (205), Expect = 9e-17, Method: Composition-based stats.
Identities = 39/93 (41%), Positives = 54/93 (58%), Gaps = 4/93 (4%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQSYLAD---RDPHYVLHSLTVQLPPSLFVENNIPVYR 58
HWG K WY +P S A + E+ ++ L D P +L L L P++ EN +PVY
Sbjct 434 HWGEAKCWYGIPGSAASAFEKVMRKTLPDLFDAQPD-LLFQLVTMLSPTVLQENKVPVYT 492
Query 59 IEQHAKEFVMLWPRTYHAGFNAGFNCNEACNFA 91
+ Q FV+ +P+++HAGFN G NC EA NFA
Sbjct 493 VLQEPGNFVITFPKSFHAGFNFGLNCAEAVNFA 525
> Hs19923370
Length=1681
Score = 79.7 bits (195), Expect = 1e-15, Method: Composition-based stats.
Identities = 39/94 (41%), Positives = 54/94 (57%), Gaps = 8/94 (8%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQS-----YLADRDPHYVLHSLTVQLPPSLFVENNIPV 56
HWG PK WY VP A +E ++ +++ D +LH L + P+ + + +PV
Sbjct 649 HWGEPKTWYGVPGYAAEQLENVMKKLAPELFVSQPD---LLHQLVTIMNPNTLMTHEVPV 705
Query 57 YRIEQHAKEFVMLWPRTYHAGFNAGFNCNEACNF 90
YR Q A EFV+ +PR YH+GFN GFN EA NF
Sbjct 706 YRTNQCAGEFVITFPRAYHSGFNQGFNFAEAVNF 739
> Hs8922311
Length=340
Score = 79.7 bits (195), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 41/92 (44%), Positives = 50/92 (54%), Gaps = 2/92 (2%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQSYL--ADRDPHYVLHSLTVQLPPSLFVENNIPVYRI 59
H G PK WYVVPP +ER + + R L + P++ EN IP RI
Sbjct 22 HLGEPKTWYVVPPEHGQRLERLARELFPGSSRGCGAFLRHKVALISPTVLKENGIPFNRI 81
Query 60 EQHAKEFVMLWPRTYHAGFNAGFNCNEACNFA 91
Q A EF++ +P YHAGFN GFNC EA NFA
Sbjct 82 TQEAGEFMVTFPYGYHAGFNHGFNCAEAINFA 113
> At5g46910
Length=707
Score = 79.0 bits (193), Expect = 2e-15, Method: Composition-based stats.
Identities = 40/100 (40%), Positives = 54/100 (54%), Gaps = 9/100 (9%)
Query 1 HHWGAPKIWYVVPPSRAPSVERTLQSYLADRD---------PHYVLHSLTVQLPPSLFVE 51
H GA K WY +P S A E+ ++ + + D VL T PP ++
Sbjct 224 QHCGASKTWYGIPGSAALKFEKVVKECVYNDDILSTNGEDGAFDVLLGKTTIFPPKTLLD 283
Query 52 NNIPVYRIEQHAKEFVMLWPRTYHAGFNAGFNCNEACNFA 91
+N+PVY+ Q EFV+ +PR YHAGF+ GFNC EA NFA
Sbjct 284 HNVPVYKAVQKPGEFVVTFPRAYHAGFSHGFNCGEAVNFA 323
> 7303354
Length=717
Score = 79.0 bits (193), Expect = 2e-15, Method: Composition-based stats.
Identities = 38/93 (40%), Positives = 56/93 (60%), Gaps = 4/93 (4%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQSYLADRDPH---YVLHSLTVQLPPSLFVENNIPVYR 58
H+GAPK WYVVPP +E+ Y + Y+ H +T+ + P + ++++PV +
Sbjct 199 HFGAPKTWYVVPPECGRKLEKVANQYFPASYKNCNAYLRHKMTL-ISPQILKQHDVPVSK 257
Query 59 IEQHAKEFVMLWPRTYHAGFNAGFNCNEACNFA 91
I Q A E ++ +P YHAGFN GFNC E+ NFA
Sbjct 258 ITQEAGEIMITFPFGYHAGFNHGFNCAESTNFA 290
> Hs17472996
Length=506
Score = 78.2 bits (191), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 40/92 (43%), Positives = 50/92 (54%), Gaps = 2/92 (2%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQSYLAD--RDPHYVLHSLTVQLPPSLFVENNIPVYRI 59
H+G PK WYVVPP +ER + D R L + P++ EN IP +
Sbjct 202 HFGEPKTWYVVPPEHGQHLERLARELFPDISRGCEAFLRHKVALISPTVLKENGIPFNCM 261
Query 60 EQHAKEFVMLWPRTYHAGFNAGFNCNEACNFA 91
Q A EF++ +P YHAGFN GFNC EA NFA
Sbjct 262 TQEAGEFMVTFPYGYHAGFNHGFNCAEAINFA 293
> Hs18573213
Length=1056
Score = 76.6 bits (187), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 37/93 (39%), Positives = 53/93 (56%), Gaps = 4/93 (4%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQSYLADRD---PHYVLHSLTVQLPPSLFVENNIPVYR 58
H+G PK WY +PP +ER Q + ++ H +T+ + PS+ + IP +
Sbjct 203 HFGEPKSWYAIPPEHGKRLERLAQGFFPSSSQGCDAFLRHKMTL-ISPSVLKKYGIPFDK 261
Query 59 IEQHAKEFVMLWPRTYHAGFNAGFNCNEACNFA 91
I Q A EF++ +P YHAGFN GFNC E+ NFA
Sbjct 262 ITQEAGEFMITFPYGYHAGFNHGFNCAESTNFA 294
> Hs11321605
Length=1560
Score = 76.3 bits (186), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 39/92 (42%), Positives = 52/92 (56%), Gaps = 4/92 (4%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQSY---LADRDPHYVLHSLTVQLPPSLFVENNIPVYR 58
HWG PK WY VP A +E ++ L D P +LH L + P+ + + +PV R
Sbjct 527 HWGEPKTWYGVPSLAAEHLEEVMKKLTPELFDSQPD-LLHQLVTLMNPNTLMSHGVPVVR 585
Query 59 IEQHAKEFVMLWPRTYHAGFNAGFNCNEACNF 90
Q A EFV+ +PR YH+GFN G+N EA NF
Sbjct 586 TNQCAGEFVITFPRAYHSGFNQGYNFAEAVNF 617
> Hs4759150
Length=1539
Score = 76.3 bits (186), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 39/92 (42%), Positives = 52/92 (56%), Gaps = 4/92 (4%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQSY---LADRDPHYVLHSLTVQLPPSLFVENNIPVYR 58
HWG PK WY VP A +E ++ L D P +LH L + P+ + + +PV R
Sbjct 517 HWGEPKTWYGVPSLAAEHLEEVMKMLTPELFDSQPD-LLHQLVTLMNPNTLMSHGVPVVR 575
Query 59 IEQHAKEFVMLWPRTYHAGFNAGFNCNEACNF 90
Q A EFV+ +PR YH+GFN G+N EA NF
Sbjct 576 TNQCAGEFVITFPRAYHSGFNQGYNFAEAVNF 607
> Hs20560402
Length=607
Score = 74.3 bits (181), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 38/92 (41%), Positives = 49/92 (53%), Gaps = 2/92 (2%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQSYL--ADRDPHYVLHSLTVQLPPSLFVENNIPVYRI 59
H+G PK WYVVPP +E + R L + P++ +N IP R+
Sbjct 202 HFGEPKTWYVVPPEHGQRLECLARELFPGNSRGCEGFLRHKVALISPTVLKKNGIPFNRM 261
Query 60 EQHAKEFVMLWPRTYHAGFNAGFNCNEACNFA 91
Q A EF++ +P YHAGFN GFNC EA NFA
Sbjct 262 TQEAGEFMVTFPYGYHAGFNHGFNCAEAINFA 293
> Hs7662246
Length=1064
Score = 74.3 bits (181), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 36/93 (38%), Positives = 53/93 (56%), Gaps = 4/93 (4%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQSYL---ADRDPHYVLHSLTVQLPPSLFVENNIPVYR 58
H+G PK WY VPP +ER + + A ++ H +T+ + P + + IP +
Sbjct 201 HFGEPKSWYSVPPEHGKRLERLAKGFFPGSAQSCEAFLRHKMTL-ISPLMLKKYGIPFDK 259
Query 59 IEQHAKEFVMLWPRTYHAGFNAGFNCNEACNFA 91
+ Q A EF++ +P YHAGFN GFNC E+ NFA
Sbjct 260 VTQEAGEFMITFPYGYHAGFNHGFNCAESTNFA 292
> Hs14764011
Length=1096
Score = 73.2 bits (178), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 36/93 (38%), Positives = 51/93 (54%), Gaps = 4/93 (4%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQSYLADRDPH---YVLHSLTVQLPPSLFVENNIPVYR 58
H+G PK WY +PP +ER + ++ H +T+ + P + + IP R
Sbjct 202 HFGEPKSWYAIPPEHGKRLERLAIGFFPGSSQGCDAFLRHKMTL-ISPIILKKYGIPFSR 260
Query 59 IEQHAKEFVMLWPRTYHAGFNAGFNCNEACNFA 91
I Q A EF++ +P YHAGFN GFNC E+ NFA
Sbjct 261 ITQEAGEFMITFPYGYHAGFNHGFNCAESTNFA 293
> SPAC1002.05c
Length=715
Score = 70.9 bits (172), Expect = 5e-13, Method: Composition-based stats.
Identities = 35/94 (37%), Positives = 51/94 (54%), Gaps = 2/94 (2%)
Query 1 HHWGAPKIWYVVPPSRAPSVERTLQSYLAD--RDPHYVLHSLTVQLPPSLFVENNIPVYR 58
H+G K+WY +P +A ER D + +L+ L + P + + VY
Sbjct 480 QHYGDTKLWYGIPGDQAERFERAALDIAPDLVKKQKDLLYQLATMINPDELQKRGVDVYF 539
Query 59 IEQHAKEFVMLWPRTYHAGFNAGFNCNEACNFAP 92
I+Q EFV+ +P+++HAG N GFN NEA NFAP
Sbjct 540 IDQGPNEFVITFPKSFHAGINHGFNINEAVNFAP 573
> 7304135
Length=495
Score = 70.9 bits (172), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 35/93 (37%), Positives = 54/93 (58%), Gaps = 4/93 (4%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQSYLADRDPH---YVLHSLTVQLPPSLFVENNIPVYR 58
H+GAPK WY +PP+ +E+ ++ Y+ H +T+ + P + ++NIP +
Sbjct 208 HFGAPKTWYAIPPAYGRRLEKLANETFSENYQECNAYLRHKMTM-ISPKVLRQHNIPYNK 266
Query 59 IEQHAKEFVMLWPRTYHAGFNAGFNCNEACNFA 91
I Q A E ++ +P YHAGFN GFN E+ NFA
Sbjct 267 ITQEAGEIMITFPFGYHAGFNHGFNGAESTNFA 299
> CE06625
Length=1430
Score = 70.5 bits (171), Expect = 6e-13, Method: Composition-based stats.
Identities = 38/97 (39%), Positives = 50/97 (51%), Gaps = 8/97 (8%)
Query 1 HHWGAPKIWYVVPPSRAPSVERTLQSYLA-----DRDPHYVLHSLTVQLPPSLFVENNIP 55
+H+G KIWY V A E L+ RD + H +T P L +P
Sbjct 479 NHFGERKIWYGVGGEDAEKFEDALKKIAPGLTGRQRD---LFHHMTTAANPHLLRSLGVP 535
Query 56 VYRIEQHAKEFVMLWPRTYHAGFNAGFNCNEACNFAP 92
++ + Q+A EFV+ +PR YHAGFN G N EA NFAP
Sbjct 536 IHSVHQNAGEFVITFPRAYHAGFNEGLNFAEAVNFAP 572
> YJR119c
Length=728
Score = 68.2 bits (165), Expect = 3e-12, Method: Composition-based stats.
Identities = 36/95 (37%), Positives = 50/95 (52%), Gaps = 6/95 (6%)
Query 1 HHWGAPKIWYVVPPSRAPSVERTLQSYLAD---RDPHYVLHSLTVQLPP--SLFVENNIP 55
H G PK+WY +P S L D + P +LH L + P F ++ IP
Sbjct 439 QHEGDPKVWYSIPESGCTKFNDLLNDMSPDLFIKQPD-LLHQLVTLISPYDPNFKKSGIP 497
Query 56 VYRIEQHAKEFVMLWPRTYHAGFNAGFNCNEACNF 90
VY+ Q E+++ +P+ YHAGFN G+N NEA NF
Sbjct 498 VYKAVQKPNEYIITFPKCYHAGFNTGYNFNEAVNF 532
> At2g38950
Length=694
Score = 64.7 bits (156), Expect = 4e-11, Method: Composition-based stats.
Identities = 32/94 (34%), Positives = 52/94 (55%), Gaps = 4/94 (4%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQSY---LADRDPHYVLHSLTVQLPPSLFVENNIPVYR 58
H GAP++WY V + ++S+ ++ P + + + P L VE IPV R
Sbjct 333 HVGAPRVWYSVAGCHRSKFKAAMKSFILEMSGEQPKKSHNPVMMMSPYQLSVEG-IPVTR 391
Query 59 IEQHAKEFVMLWPRTYHAGFNAGFNCNEACNFAP 92
QH ++V+++P +Y++ F+ GFNC E NFAP
Sbjct 392 CVQHPGQYVIIFPGSYYSAFDCGFNCLEKANFAP 425
> SPAC343.11c
Length=1588
Score = 63.5 bits (153), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 33/95 (34%), Positives = 54/95 (56%), Gaps = 6/95 (6%)
Query 1 HHWGAPKIWYVVPPSRAPSVERTLQ----SYLADRDPHYVLHSLTVQLPPSLFVENNIPV 56
HH+GA +IWYV+P P E+ L S++ ++ P ++ S + LP S+ + N I V
Sbjct 537 HHYGAQRIWYVIPEVDGPKYEKLLNDLSPSFIQEK-PETLIKS-KILLPISMLISNGIQV 594
Query 57 YRIEQHAKEFVMLWPRTYHAGFNAGFNCNEACNFA 91
Q++ EFV+ P TY+ + GF+ +E+ FA
Sbjct 595 LTFVQNSNEFVITSPNTYYTVLDTGFSLSESVPFA 629
> Hs20477063
Length=1011
Score = 63.2 bits (152), Expect = 1e-10, Method: Composition-based stats.
Identities = 27/55 (49%), Positives = 36/55 (65%), Gaps = 0/55 (0%)
Query 36 VLHSLTVQLPPSLFVENNIPVYRIEQHAKEFVMLWPRTYHAGFNAGFNCNEACNF 90
+LH L + P+ + + +PVYR Q A EFV+ +PR YH+GFN GFN EA NF
Sbjct 15 LLHQLVTIMNPNTLMTHEVPVYRTNQCAGEFVITFPRAYHSGFNQGFNFAEAVNF 69
> SPBP19A11.06
Length=1513
Score = 61.2 bits (147), Expect = 5e-10, Method: Composition-based stats.
Identities = 33/97 (34%), Positives = 48/97 (49%), Gaps = 10/97 (10%)
Query 1 HHWGAPKIWYVVPPSRAPSVERTLQSYLADRDPHYVLHSLT------VQLPPSLFVENNI 54
H +G WYV+PP + + ER YL P Y + L V + PS +EN
Sbjct 466 HRFGDTVTWYVLPPDESDAFER----YLISSYPQYTMEDLNRSNGLPVIVSPSSLIENGF 521
Query 55 PVYRIEQHAKEFVMLWPRTYHAGFNAGFNCNEACNFA 91
I+ EF+++ P +YH GF+ GF+ E+ NFA
Sbjct 522 HPIAIDLRPNEFLVVSPNSYHMGFHQGFSSFESVNFA 558
> YER169w_1
Length=706
Score = 60.8 bits (146), Expect = 6e-10, Method: Composition-based stats.
Identities = 34/93 (36%), Positives = 47/93 (50%), Gaps = 4/93 (4%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQSYL---ADRDPHYVLHSLTVQLPPSLFVENNIPVYR 58
H+GAPK WY +P + +Q A P ++ H + + P L EN I
Sbjct 248 HFGAPKQWYSIPQEDRFKFYKFMQEQFPEEAKNCPEFLRHKMFLA-SPKLLQENGIRCNE 306
Query 59 IEQHAKEFVMLWPRTYHAGFNAGFNCNEACNFA 91
I H EF++ +P YHAGFN G+N E+ NFA
Sbjct 307 IVHHEGEFMITYPYGYHAGFNYGYNLAESVNFA 339
> At1g30810
Length=787
Score = 60.5 bits (145), Expect = 7e-10, Method: Composition-based stats.
Identities = 34/92 (36%), Positives = 45/92 (48%), Gaps = 30/92 (32%)
Query 1 HHWGAPKIWYVVPPSRAPSVERTLQSYLADRDPHYVLHSLTVQLPPSLFVENNIPVYRIE 60
HH+G PK+WY VP S A ++E+ ++ +L P LF ++ I
Sbjct 319 HHFGEPKVWYGVPGSNATALEKAMRKHL-----------------PDLFEDSLI------ 355
Query 61 QHAKEFVMLWPRTYHAGFNAGFNCNEACNFAP 92
+ M W YHAGFN GFNC EA N AP
Sbjct 356 -----YYMAW--AYHAGFNCGFNCAEAVNVAP 380
> CE22126
Length=894
Score = 59.7 bits (143), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 34/95 (35%), Positives = 47/95 (49%), Gaps = 5/95 (5%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQ---SYLADRDPH--YVLHSLTVQLPPSLFVENNIPV 56
H+GAPK W+ + A ER + SY + P L T + P L + IP
Sbjct 278 HFGAPKYWFAISSEHADRFERFMSQQFSYQNEYAPQCKAFLRHKTYLVTPELLRQAGIPY 337
Query 57 YRIEQHAKEFVMLWPRTYHAGFNAGFNCNEACNFA 91
+ Q EF++ +PR YH GFN G+N E+ NFA
Sbjct 338 ATMVQRPNEFIITFPRGYHMGFNLGYNLAESTNFA 372
> At5g04240_1
Length=1165
Score = 55.8 bits (133), Expect = 2e-08, Method: Composition-based stats.
Identities = 31/83 (37%), Positives = 44/83 (53%), Gaps = 5/83 (6%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQ--SYLADRDPHYVLHSL---TVQLPPSLFVENNIPV 56
H G+PK WY VP A E ++ SY + D L L T + P + V + IP
Sbjct 318 HTGSPKTWYAVPCDYALDFEEVIRKNSYGRNIDQLAALTQLGEKTTLVSPEMIVASGIPC 377
Query 57 YRIEQHAKEFVMLWPRTYHAGFN 79
R+ Q+ EFV+ +PR+YH GF+
Sbjct 378 CRLVQNPGEFVVTFPRSYHVGFS 400
> YDR096w_1
Length=734
Score = 52.0 bits (123), Expect = 3e-07, Method: Composition-based stats.
Identities = 28/92 (30%), Positives = 45/92 (48%), Gaps = 4/92 (4%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQ---SYLADRDPHYVLHSLTVQLPPSLFVENNIPVYR 58
H+GAPK WY +P + + L S + P ++ H + P +NNI R
Sbjct 217 HFGAPKQWYSIPSANTDQFLKILSKEPSSNKENCPAFIRHQ-NIITSPDFLRKNNIKFNR 275
Query 59 IEQHAKEFVMLWPRTYHAGFNAGFNCNEACNF 90
+ Q EF++ +P ++GFN G+N E+ F
Sbjct 276 VVQFQHEFIITFPYCMYSGFNYGYNFGESIEF 307
> Hs20483920
Length=114
Score = 45.4 bits (106), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 26/91 (28%), Positives = 41/91 (45%), Gaps = 15/91 (16%)
Query 1 HHWGAPKIWYVVPPSRAPSVERTLQSYLADRDPHYVLHSLTVQLPPSLFVENNIPVYRIE 60
HH+ P WY VP + + +E +++ + P++ ++N IP +
Sbjct 8 HHFREPLYWYAVPQNHSWHLECSVRELFQ-----------MALIVPTVLMQNGIPFNSMS 56
Query 61 QHAKEFVMLWPRTYHAGFNAGFNCNEACNFA 91
Q A EF + +P Y N GFNC EA N
Sbjct 57 QEAGEFTVTFPYDY----NTGFNCKEAINLT 83
> Hs11863152
Length=1246
Score = 44.7 bits (104), Expect = 4e-05, Method: Composition-based stats.
Identities = 24/91 (26%), Positives = 45/91 (49%), Gaps = 1/91 (1%)
Query 2 HWGAPKIWYVVPPSRAPSVERTLQSYL-ADRDPHYVLHSLTVQLPPSLFVENNIPVYRIE 60
H GA IWY +P +E + + L A+ P + V + P + + I V+R
Sbjct 942 HTGADCIWYCIPAEEENKLEDVVHTLLQANGTPGLQMLESNVMISPEVLCKEGIKVHRTV 1001
Query 61 QHAKEFVMLWPRTYHAGFNAGFNCNEACNFA 91
Q + +FV+ +P ++ + G++ +E +FA
Sbjct 1002 QQSGQFVVCFPGSFVSKVCCGYSVSETVHFA 1032
> SPBC83.07
Length=752
Score = 41.6 bits (96), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/84 (28%), Positives = 39/84 (46%), Gaps = 4/84 (4%)
Query 1 HHWGAPKIWYVVPPSRAPSVERTLQSYLAD---RDPHYVLHSLTVQLPPSLFVENNIPVY 57
H G+ WYV+P + + S ++ D R ++LH + PPS V+N I Y
Sbjct 225 HLGGSSLQWYVIPSAHSESFKKLAGKLAQDEHWRCSDFLLHQ-NILFPPSTLVQNGIVTY 283
Query 58 RIEQHAKEFVMLWPRTYHAGFNAG 81
E ++ +P T+H+ F G
Sbjct 284 STVLKQDELLITFPGTHHSAFCLG 307
> YMR176w
Length=1411
Score = 32.3 bits (72), Expect = 0.19, Method: Composition-based stats.
Identities = 11/47 (23%), Positives = 23/47 (48%), Gaps = 0/47 (0%)
Query 46 PSLFVENNIPVYRIEQHAKEFVMLWPRTYHAGFNAGFNCNEACNFAP 92
P+ + N I +Y+ Q ++ +P+ + +GF ++ FAP
Sbjct 634 PNFILANGIKLYKTTQEQGSYIFKFPKAFTCSIGSGFYLSQNAKFAP 680
> CE25300
Length=234
Score = 30.4 bits (67), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 18/55 (32%), Positives = 27/55 (49%), Gaps = 0/55 (0%)
Query 14 PSRAPSVERTLQSYLADRDPHYVLHSLTVQLPPSLFVENNIPVYRIEQHAKEFVM 68
PS P+VE + YLA D + +L ++ VQ F + I V +AK V+
Sbjct 29 PSPIPNVEEGAKLYLASNDDNGLLKNIQVQTAGGSFTLDQINVNTANGNAKSIVI 83
> At1g02080
Length=2226
Score = 29.6 bits (65), Expect = 1.2, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 21/43 (48%), Gaps = 6/43 (13%)
Query 42 VQLPPS------LFVENNIPVYRIEQHAKEFVMLWPRTYHAGF 78
++ PPS F+ NNI IE KEF + P+ Y+ F
Sbjct 812 IEAPPSDVQDKVSFIINNISTTNIESKGKEFAEILPQQYYPWF 854
> CE16630
Length=595
Score = 27.3 bits (59), Expect = 7.3, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 22/40 (55%), Gaps = 1/40 (2%)
Query 43 QLPPSLFVENNIPVYRIEQHAKEFVMLWPRTYHAGFNAGF 82
+L ++FV NNIPVY + + V+ W T G +AG
Sbjct 108 ELSANVFVRNNIPVYLFSEVSPTPVVSWA-TIKLGCDAGL 146
> 7303706
Length=480
Score = 27.3 bits (59), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 20/38 (52%), Gaps = 5/38 (13%)
Query 30 DRDPHYVLHSLTVQLPP-SLFVENNIPVYRIEQHAKEF 66
DR PH + +LPP L VEN I +QHAK+
Sbjct 33 DRSPH----NHNAKLPPLELLVENKISYSSYQQHAKDL 66
Lambda K H
0.323 0.137 0.473
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171209254
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40