bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1549_orf1
Length=137
Score E
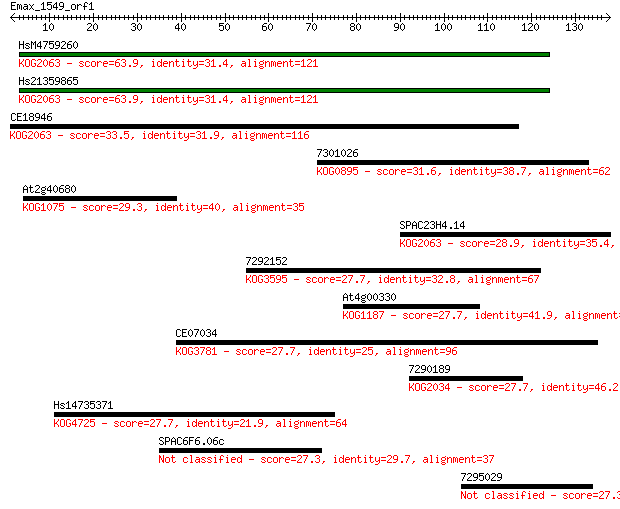
Sequences producing significant alignments: (Bits) Value
HsM4759260 63.9 8e-11
Hs21359865 63.9 8e-11
CE18946 33.5 0.11
7301026 31.6 0.47
At2g40680 29.3 2.1
SPAC23H4.14 28.9 3.2
7292152 27.7 5.8
At4g00330 27.7 6.6
CE07034 27.7 6.8
7290189 27.7 7.4
Hs14735371 27.7 7.4
SPAC6F6.06c 27.3 8.5
7295029 27.3 9.6
> HsM4759260
Length=860
Score = 63.9 bits (154), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 38/122 (31%), Positives = 67/122 (54%), Gaps = 2/122 (1%)
Query 3 SAVDLWRKLAKGELQDPHYSG-XDRVVNLLKNCQNSRLILNNIEFLLEHNQEMAVEILIL 61
+AV LW + G++QD S + +V+ L C + L+ +++L+ ++E+ V++
Sbjct 504 AAVQLWVNIVNGDVQDSTRSDLYEYIVDFLTYCLDEELVWAYADWVLQKSEEVGVQVFTK 563
Query 62 NTRMEDNKFPVLEPEHVITVLHKYRRALIIYLEFVVFDLKVKSTPIHNRLAMLYVEIISL 121
E K P+ +I L KY +AL+ YLE +V D +++ H LA+LY+E + L
Sbjct 564 RPLDEQQK-NSFNPDDIINCLKKYPKALVKYLEHLVIDKRLQKEEYHTHLAVLYLEEVLL 622
Query 122 QR 123
QR
Sbjct 623 QR 624
> Hs21359865
Length=860
Score = 63.9 bits (154), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 38/122 (31%), Positives = 67/122 (54%), Gaps = 2/122 (1%)
Query 3 SAVDLWRKLAKGELQDPHYSG-XDRVVNLLKNCQNSRLILNNIEFLLEHNQEMAVEILIL 61
+AV LW + G++QD S + +V+ L C + L+ +++L+ ++E+ V++
Sbjct 504 AAVQLWVNIVNGDVQDSTRSDLYEYIVDFLTYCLDEELVWAYADWVLQKSEEVGVQVFTK 563
Query 62 NTRMEDNKFPVLEPEHVITVLHKYRRALIIYLEFVVFDLKVKSTPIHNRLAMLYVEIISL 121
E K P+ +I L KY +AL+ YLE +V D +++ H LA+LY+E + L
Sbjct 564 RPLDEQQK-NSFNPDDIINCLKKYPKALVKYLEHLVIDKRLQKEEYHTHLAVLYLEEVLL 622
Query 122 QR 123
QR
Sbjct 623 QR 624
> CE18946
Length=923
Score = 33.5 bits (75), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 37/121 (30%), Positives = 57/121 (47%), Gaps = 9/121 (7%)
Query 1 YESAVDLWRKLAKGELQDPHYS-GXDRVVNLLKNCQNSRL--ILNNIEFLLEHNQEMAVE 57
+E A+DL+ + DP + ++V L++ NS L IL +++L N E V+
Sbjct 526 HEMALDLFIDQSSRPDADPFFDDAIQQIVEYLQSLGNSNLPLILKYAKWVLAKNLEAGVQ 585
Query 58 ILILN-TRMEDNKFPVLEPEHVITVL-HKYRRALIIYLEFVVFDLKVKSTPIHNRLAMLY 115
I + T M N L + V+ L + ALI YLE V+F + S+ H L Y
Sbjct 586 IFTSDETEMARN----LNRKAVVEFLKSECPDALIPYLEHVIFKWEEPSSYFHETLLEFY 641
Query 116 V 116
V
Sbjct 642 V 642
> 7301026
Length=1398
Score = 31.6 bits (70), Expect = 0.47, Method: Composition-based stats.
Identities = 24/75 (32%), Positives = 34/75 (45%), Gaps = 13/75 (17%)
Query 71 PVLEPEHVIT--VLHKYRRALI-----------IYLEFVVFDLKVKSTPIHNRLAMLYVE 117
P+ EP +I LHK +R + YL++ DL VK N A Y+
Sbjct 331 PLKEPRSLIKGEELHKVKRLSLYESYELQIHDRFYLKYSKCDLLVKYADWENEQAEKYIP 390
Query 118 IISLQREMEEKDSKL 132
I Q++M+ K SKL
Sbjct 391 IFHAQKKMQRKASKL 405
> At2g40680
Length=296
Score = 29.3 bits (64), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 19/35 (54%), Gaps = 0/35 (0%)
Query 4 AVDLWRKLAKGELQDPHYSGXDRVVNLLKNCQNSR 38
+ +W KL KG LQ + S DR+V LL + R
Sbjct 190 STQVWEKLMKGLLQGHYTSSWDRIVTLLTSSSLGR 224
> SPAC23H4.14
Length=905
Score = 28.9 bits (63), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 31/49 (63%), Gaps = 1/49 (2%)
Query 90 IIYLEFVVFDLKVKSTPIHNRLAMLYVE-IISLQREMEEKDSKLCEKTV 137
IIYLE ++ D K T RLA+LY++ I+ L+ + K+ ++ ++T+
Sbjct 641 IIYLEKLLLDNKFNDTVFPTRLALLYLKRILELEETTDFKNQEVFKQTI 689
> 7292152
Length=3868
Score = 27.7 bits (60), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 34/67 (50%), Gaps = 6/67 (8%)
Query 55 AVEILILNTRMEDNKFPVLEPEHVITVLHKYRRALIIYLEFVVFDLKVKSTPIHNRLAML 114
AV I +NT ++DNK L VIT+ ++ + + F V DL S +R M+
Sbjct 1649 AVWIENMNTVLDDNKKLCLTSGEVITMSNE------MSMVFEVMDLAQASPATVSRCGMI 1702
Query 115 YVEIISL 121
Y+E +L
Sbjct 1703 YMEPSTL 1709
> At4g00330
Length=341
Score = 27.7 bits (60), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 77 HVITVLHKYRRALIIYLEFVVFDLKVKSTPI 107
H IT LH Y R I+Y+E + +KS+ I
Sbjct 153 HAITYLHMYTRKSIVYIEPPIIHRDIKSSNI 183
> CE07034
Length=331
Score = 27.7 bits (60), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 24/101 (23%), Positives = 48/101 (47%), Gaps = 7/101 (6%)
Query 39 LILNNIEFLLEH-NQEMAVEILILNTRMEDNKFPVLEPEHVITVLHKYRRALIIYLEFVV 97
L+ E+L+ + N E+ E+ + ++F ++ + + + R +I+LEF+
Sbjct 29 LVAGTSEYLVNNKNMEILFEVHVRENVTPKHRFSLVLLRPTVEQMLGFPRTRVIFLEFLA 88
Query 98 FDLKVKSTPIHN----RLAMLYVEIISLQREMEEKDSKLCE 134
+ + S I N RL+ + I+S KDS+LC+
Sbjct 89 NAMNISSISILNIEYIRLSPDNMTIVSFHN--NSKDSELCD 127
> 7290189
Length=1002
Score = 27.7 bits (60), Expect = 7.4, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 15/26 (57%), Gaps = 0/26 (0%)
Query 92 YLEFVVFDLKVKSTPIHNRLAMLYVE 117
YLEF ++ L + IHN L LY E
Sbjct 681 YLEFAIYKLNTTNDAIHNFLLHLYAE 706
> Hs14735371
Length=834
Score = 27.7 bits (60), Expect = 7.4, Method: Composition-based stats.
Identities = 14/64 (21%), Positives = 33/64 (51%), Gaps = 0/64 (0%)
Query 11 LAKGELQDPHYSGXDRVVNLLKNCQNSRLILNNIEFLLEHNQEMAVEILILNTRMEDNKF 70
+ K E H +G ++ + + +I++NI+ +++ N+ + EIL + R+E+
Sbjct 149 MTKVEELQKHSAGNSMLIPSMSVTMETSMIMSNIQRIIQENERLKQEILEKSNRIEEQND 208
Query 71 PVLE 74
+ E
Sbjct 209 KISE 212
> SPAC6F6.06c
Length=1155
Score = 27.3 bits (59), Expect = 8.5, Method: Composition-based stats.
Identities = 11/37 (29%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 35 QNSRLILNNIEFLLEHNQEMAVEILILNTRMEDNKFP 71
+NSR++ ++I FL + ++ ++ +NT + NK P
Sbjct 492 KNSRMVTHSIRFLEQSYTNVSNGLVFVNTTTDVNKLP 528
> 7295029
Length=1274
Score = 27.3 bits (59), Expect = 9.6, Method: Compositional matrix adjust.
Identities = 11/30 (36%), Positives = 18/30 (60%), Gaps = 0/30 (0%)
Query 104 STPIHNRLAMLYVEIISLQREMEEKDSKLC 133
STP+H A ++I+ L EM+ + +LC
Sbjct 320 STPVHLACAQGAIDIVKLMFEMQPMEKRLC 349
Lambda K H
0.322 0.138 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1461889840
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40