bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1565_orf1
Length=88
Score E
Sequences producing significant alignments: (Bits) Value
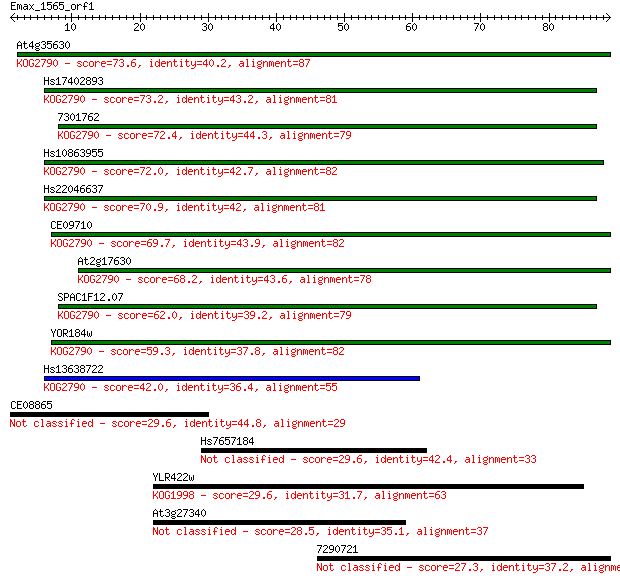
At4g35630 73.6 9e-14
Hs17402893 73.2 1e-13
7301762 72.4 2e-13
Hs10863955 72.0 2e-13
Hs22046637 70.9 6e-13
CE09710 69.7 1e-12
At2g17630 68.2 4e-12
SPAC1F12.07 62.0 3e-10
YOR184w 59.3 2e-09
Hs13638722 42.0 3e-04
CE08865 29.6 1.3
Hs7657184 29.6 1.3
YLR422w 29.6 1.5
At3g27340 28.5 3.6
7290721 27.3 6.8
> At4g35630
Length=430
Score = 73.6 bits (179), Expect = 9e-14, Method: Composition-based stats.
Identities = 35/87 (40%), Positives = 52/87 (59%), Gaps = 6/87 (6%)
Query 2 IQPFTDWTLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTKQIP 61
+ F + +A YLH C NETIHG+EF +P +P +LVAD +SN +K +
Sbjct 199 VPSFEELEQTPDAKYLHICANETIHGVEFKDYP------VPKNGFLVADMSSNFCSKPVD 252
Query 62 WNIIDLAYAGAQKNIGIAGVTLVVVHK 88
+ + Y GAQKN+G +GVT+V++ K
Sbjct 253 VSKFGVIYGGAQKNVGPSGVTIVIIRK 279
> Hs17402893
Length=370
Score = 73.2 bits (178), Expect = 1e-13, Method: Composition-based stats.
Identities = 35/81 (43%), Positives = 51/81 (62%), Gaps = 6/81 (7%)
Query 6 TDWTLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTKQIPWNII 65
+ W LN +A+Y++YC NET+HG+EF PD LV D +SN +K + +
Sbjct 138 STWNLNPDASYVYYCANETVHGVEFDFIPDVKG------AVLVCDMSSNFLSKPVDVSKF 191
Query 66 DLAYAGAQKNIGIAGVTLVVV 86
+ +AGAQKN+G AGVT+V+V
Sbjct 192 GVIFAGAQKNVGSAGVTVVIV 212
> 7301762
Length=364
Score = 72.4 bits (176), Expect = 2e-13, Method: Composition-based stats.
Identities = 35/79 (44%), Positives = 52/79 (65%), Gaps = 5/79 (6%)
Query 8 WTLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTKQIPWNIIDL 67
W L+ NA+Y++YCDNET+ G+EF P+ +P+ LVAD +SN ++ + +
Sbjct 133 WKLDPNASYVYYCDNETVEGVEFDFVPE-----VPAGVPLVADMSSNFLSRPFDVSKFGV 187
Query 68 AYAGAQKNIGIAGVTLVVV 86
YAGAQKNIG AG T+++V
Sbjct 188 IYAGAQKNIGPAGTTVIIV 206
> Hs10863955
Length=324
Score = 72.0 bits (175), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 35/82 (42%), Positives = 51/82 (62%), Gaps = 6/82 (7%)
Query 6 TDWTLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTKQIPWNII 65
+ W LN +A+Y++YC NET+HG+EF PD LV D +SN +K + +
Sbjct 138 STWNLNPDASYVYYCANETVHGVEFDFIPDVKG------AVLVCDMSSNFLSKPVDVSKF 191
Query 66 DLAYAGAQKNIGIAGVTLVVVH 87
+ +AGAQKN+G AGVT+V+V
Sbjct 192 GVIFAGAQKNVGSAGVTVVIVR 213
> Hs22046637
Length=481
Score = 70.9 bits (172), Expect = 6e-13, Method: Composition-based stats.
Identities = 34/81 (41%), Positives = 49/81 (60%), Gaps = 6/81 (7%)
Query 6 TDWTLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTKQIPWNII 65
+ W LN A+Y++YC N T+HG+EF PD LV D SN +K + +
Sbjct 149 STWNLNPEASYVYYCANGTVHGVEFDFIPDVKG------AVLVCDMFSNFLSKPVDVSKF 202
Query 66 DLAYAGAQKNIGIAGVTLVVV 86
D+ ++GAQKN+G AGVT+V+V
Sbjct 203 DVIFSGAQKNVGSAGVTVVIV 223
> CE09710
Length=370
Score = 69.7 bits (169), Expect = 1e-12, Method: Composition-based stats.
Identities = 36/83 (43%), Positives = 55/83 (66%), Gaps = 7/83 (8%)
Query 7 DWTLNKNAAYLHYCDNETIHGLEF-PSFPDFSSFSLPSPRYLVADFASNLGTKQIPWNII 65
+W ++ AAYL+YC NET+HG+EF P+ P+ S ++P LVAD +SN + +
Sbjct 137 NWVHDEKAAYLYYCANETVHGIEFTPTAPE--SHNVP----LVADVSSNFMARPFDFKDH 190
Query 66 DLAYAGAQKNIGIAGVTLVVVHK 88
+ + GAQKN+G AG+T+V+V K
Sbjct 191 GVVFGGAQKNLGAAGLTIVIVRK 213
> At2g17630
Length=422
Score = 68.2 bits (165), Expect = 4e-12, Method: Composition-based stats.
Identities = 34/79 (43%), Positives = 51/79 (64%), Gaps = 6/79 (7%)
Query 11 NKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPR-YLVADFASNLGTKQIPWNIIDLAY 69
+ +A YLH C NETIHG+EF +P + +P L+AD +SN +K + + + Y
Sbjct 198 SSDAKYLHICANETIHGVEFKDYP-----LVENPDGVLIADMSSNFCSKPVDVSKFGVIY 252
Query 70 AGAQKNIGIAGVTLVVVHK 88
AGAQKN+G +GVT+V++ K
Sbjct 253 AGAQKNVGPSGVTIVIIRK 271
> SPAC1F12.07
Length=389
Score = 62.0 bits (149), Expect = 3e-10, Method: Composition-based stats.
Identities = 31/79 (39%), Positives = 49/79 (62%), Gaps = 4/79 (5%)
Query 8 WTLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTKQIPWNIIDL 67
+T + + ++YCDNET+HG+EF P ++P V D +SN +++I + D+
Sbjct 153 FTPDGETSLVYYCDNETVHGVEFNEPPT----NIPKGAIRVCDVSSNFISRKIDFTKHDI 208
Query 68 AYAGAQKNIGIAGVTLVVV 86
+AGAQKN G AG+T+V V
Sbjct 209 IFAGAQKNAGPAGITVVFV 227
> YOR184w
Length=395
Score = 59.3 bits (142), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 31/83 (37%), Positives = 52/83 (62%), Gaps = 4/83 (4%)
Query 7 DWTLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPRY-LVADFASNLGTKQIPWNII 65
D K +Y++ C+NET+HG+E+P P + P +VAD +S++ +++I +
Sbjct 153 DKIKGKAFSYVYLCENETVHGVEWPELP---KCLVNDPNIEIVADLSSDILSRKIDVSQY 209
Query 66 DLAYAGAQKNIGIAGVTLVVVHK 88
+ AGAQKNIG+AG+TL ++ K
Sbjct 210 GVIMAGAQKNIGLAGLTLYIIKK 232
> Hs13638722
Length=212
Score = 42.0 bits (97), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 30/55 (54%), Gaps = 6/55 (10%)
Query 6 TDWTLNKNAAYLHYCDNETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTKQI 60
+ W LN +A+Y++YC NE +HG+EF P LV D +SN +K +
Sbjct 138 STWNLNPDASYVYYCANEVVHGVEFDFIPYVKG------AVLVCDMSSNFLSKPV 186
> CE08865
Length=893
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 13/29 (44%), Positives = 17/29 (58%), Gaps = 0/29 (0%)
Query 1 DIQPFTDWTLNKNAAYLHYCDNETIHGLE 29
D+ PF ++LNK + CD ETI LE
Sbjct 674 DLTPFVKYSLNKKRRWETKCDLETISQLE 702
> Hs7657184
Length=2099
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 18/33 (54%), Gaps = 1/33 (3%)
Query 29 EFPSFPDFSSFSLPSPRYLVADFASNLGTKQIP 61
+F P FS F L SP+ LV DF + G + P
Sbjct 1881 DFEPIPSFSGFPLDSPKTLVLDFETE-GERNSP 1912
> YLR422w
Length=1932
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 20/67 (29%), Positives = 30/67 (44%), Gaps = 6/67 (8%)
Query 22 NETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTKQIPWNII----DLAYAGAQKNIG 77
N+T+ + FP P+ SSF YL+ + A N Q P + DL A + N
Sbjct 1455 NDTLEAISFPPLPEQSSFE--RKEYLLKESARNFSRGQKPEKALAVYKDLIKAYDEINYD 1512
Query 78 IAGVTLV 84
+ G+ V
Sbjct 1513 LNGLAFV 1519
> At3g27340
Length=141
Score = 28.5 bits (62), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 13/37 (35%), Positives = 21/37 (56%), Gaps = 2/37 (5%)
Query 22 NETIHGLEFPSFPDFSSFSLPSPRYLVADFASNLGTK 58
+ +HG F + P S FS+ SP+ + ++A GTK
Sbjct 9 SRRVHGTSFITIPKLSGFSIVSPKQVEVEYAD--GTK 43
> 7290721
Length=394
Score = 27.3 bits (59), Expect = 6.8, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 28/47 (59%), Gaps = 5/47 (10%)
Query 46 YLVADFASNLGTKQI---PWNIIDLAYAGAQKNI-GIAGVTLVVVHK 88
+L+ D ++LG + W + D+AY G+QK++ G AG+T + K
Sbjct 175 FLIVDTVASLGGTEFLMDEWKV-DVAYTGSQKSLGGPAGLTPISFSK 220
Lambda K H
0.321 0.138 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1184307974
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40