bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1611_orf1
Length=105
Score E
Sequences producing significant alignments: (Bits) Value
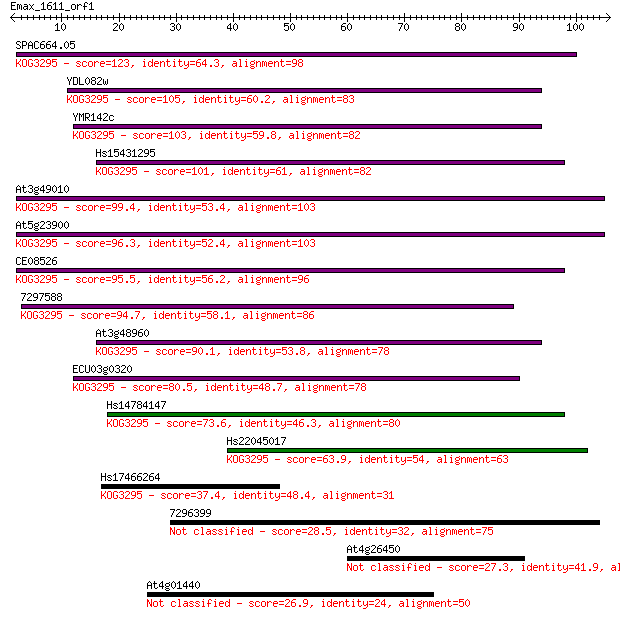
SPAC664.05 123 8e-29
YDL082w 105 2e-23
YMR142c 103 6e-23
Hs15431295 101 3e-22
At3g49010 99.4 2e-21
At5g23900 96.3 1e-20
CE08526 95.5 2e-20
7297588 94.7 4e-20
At3g48960 90.1 8e-19
ECU03g0320 80.5 7e-16
Hs14784147 73.6 8e-14
Hs22045017 63.9 7e-11
Hs17466264 37.4 0.008
7296399 28.5 3.3
At4g26450 27.3 7.6
At4g01440 26.9 9.2
> SPAC664.05
Length=208
Score = 123 bits (309), Expect = 8e-29, Method: Compositional matrix adjust.
Identities = 63/99 (63%), Positives = 76/99 (76%), Gaps = 1/99 (1%)
Query 2 QKKAAINGVRPVDLLRPAVRCPTQKYNIKVREGRGFTLEELKAVGVSPRVALSIGIAVDH 61
Q KAA RPV+ +RPAV+ PT +YN+KVR GRGFTLEELKA GVS RVA +IGI VDH
Sbjct 40 QTKAAKIAPRPVEAIRPAVKPPTIRYNMKVRAGRGFTLEELKAAGVSRRVASTIGIPVDH 99
Query 62 RRRNRSQESLNNNINRLKLYLNKLVLFQR-GSKPKKGLA 99
RRRNRS+ESL N+ R+K+YL L++F R +PKKG A
Sbjct 100 RRRNRSEESLQRNVERIKVYLAHLIVFPRKAGQPKKGDA 138
> YDL082w
Length=199
Score = 105 bits (262), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 50/83 (60%), Positives = 63/83 (75%), Gaps = 0/83 (0%)
Query 11 RPVDLLRPAVRCPTQKYNIKVREGRGFTLEELKAVGVSPRVALSIGIAVDHRRRNRSQES 70
RP+DLLRP VR PT KYN KVR GRGFTL E+KA G++ A +IGIAVDHRR+NR+QE
Sbjct 49 RPLDLLRPVVRAPTVKYNRKVRAGRGFTLAEVKAAGLTAAYARTIGIAVDHRRQNRNQEI 108
Query 71 LNNNINRLKLYLNKLVLFQRGSK 93
+ N+ RLK Y +K+++F R K
Sbjct 109 FDANVQRLKEYQSKIIVFPRNGK 131
> YMR142c
Length=199
Score = 103 bits (258), Expect = 6e-23, Method: Compositional matrix adjust.
Identities = 49/82 (59%), Positives = 62/82 (75%), Gaps = 0/82 (0%)
Query 12 PVDLLRPAVRCPTQKYNIKVREGRGFTLEELKAVGVSPRVALSIGIAVDHRRRNRSQESL 71
P+DLLRP VR PT KYN KVR GRGFTL E+KA G++ A +IGIAVDHRR+NR+QE
Sbjct 50 PLDLLRPVVRAPTVKYNRKVRAGRGFTLAEVKAAGLTAAYARTIGIAVDHRRQNRNQEIF 109
Query 72 NNNINRLKLYLNKLVLFQRGSK 93
+ N+ RLK Y +K+++F R K
Sbjct 110 DANVQRLKEYQSKIIVFPRDGK 131
> Hs15431295
Length=211
Score = 101 bits (252), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 50/83 (60%), Positives = 63/83 (75%), Gaps = 1/83 (1%)
Query 16 LRPAVRCPTQKYNIKVREGRGFTLEELKAVGVSPRVALSIGIAVDHRRRNRSQESLNNNI 75
+RP VRCPT +Y+ KVR GRGF+LEEL+ G+ +VA +IGI+VD RRRN+S ESL N+
Sbjct 55 IRPIVRCPTVRYHTKVRAGRGFSLEELRVAGIHKKVARTIGISVDPRRRNKSTESLQANV 114
Query 76 NRLKLYLNKLVLFQRG-SKPKKG 97
RLK Y +KL+LF R S PKKG
Sbjct 115 QRLKEYRSKLILFPRKPSAPKKG 137
> At3g49010
Length=206
Score = 99.4 bits (246), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 55/104 (52%), Positives = 67/104 (64%), Gaps = 1/104 (0%)
Query 2 QKKAAINGVRPVD-LLRPAVRCPTQKYNIKVREGRGFTLEELKAVGVSPRVALSIGIAVD 60
QKKA RP LRP V T KYN+KVR G+GFTLEELKA G+ ++A +IGIAVD
Sbjct 38 QKKAVKIFPRPTSGPLRPVVHGQTLKYNMKVRTGKGFTLEELKAAGIPKKLAPTIGIAVD 97
Query 61 HRRRNRSQESLNNNINRLKLYLNKLVLFQRGSKPKKGLAGVPAD 104
HRR+NRS E L N+ RLK Y KLV+F R ++ K P +
Sbjct 98 HRRKNRSLEGLQTNVQRLKTYKTKLVIFPRRARKVKAGDSTPEE 141
> At5g23900
Length=206
Score = 96.3 bits (238), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 54/104 (51%), Positives = 67/104 (64%), Gaps = 1/104 (0%)
Query 2 QKKAAINGVRPVD-LLRPAVRCPTQKYNIKVREGRGFTLEELKAVGVSPRVALSIGIAVD 60
QKKA RP LRP V T KYN+KVR G+GFTLEELK G+ ++A +IGI+VD
Sbjct 38 QKKAVKIFPRPTSGPLRPVVHGQTLKYNMKVRAGKGFTLEELKVAGIPKKLAPTIGISVD 97
Query 61 HRRRNRSQESLNNNINRLKLYLNKLVLFQRGSKPKKGLAGVPAD 104
HRR+NRS E L +N+ RLK Y KLV+F R S+ K P +
Sbjct 98 HRRKNRSLEGLQSNVQRLKTYKAKLVVFPRRSRQVKAGDSTPEE 141
> CE08526
Length=207
Score = 95.5 bits (236), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 54/98 (55%), Positives = 66/98 (67%), Gaps = 2/98 (2%)
Query 2 QKKAAINGVRPV-DLLRPAVRCPTQKYNIKVREGRGFTLEELKAVGVSPRVALSIGIAVD 60
Q KA RPV LLR VRCP ++YN K R GRGF+L+ELKA G+S A +IGIAVD
Sbjct 40 QAKAVEIAPRPVAGLLRSVVRCPQKRYNTKTRLGRGFSLQELKAAGISQAQARTIGIAVD 99
Query 61 HRRRNRSQESLNNNINRLKLYLNKLVLF-QRGSKPKKG 97
RR N++ E L N +RLK Y KL+LF ++ S PKKG
Sbjct 100 VRRTNKTAEGLKANADRLKEYKAKLILFPKKASAPKKG 137
> 7297588
Length=218
Score = 94.7 bits (234), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 50/87 (57%), Positives = 59/87 (67%), Gaps = 1/87 (1%)
Query 3 KKAAINGVRPVD-LLRPAVRCPTQKYNIKVREGRGFTLEELKAVGVSPRVALSIGIAVDH 61
KKA RP LRP VRCPT +Y+ K+R GRGFTLEELK G+ A +IGIAVD
Sbjct 40 KKAKAVFPRPASGALRPVVRCPTIRYHTKLRAGRGFTLEELKGAGIGANFAKTIGIAVDR 99
Query 62 RRRNRSQESLNNNINRLKLYLNKLVLF 88
RR+N+S ES NI RLK Y +KL+LF
Sbjct 100 RRKNKSLESRQRNIQRLKEYRSKLILF 126
> At3g48960
Length=206
Score = 90.1 bits (222), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 42/78 (53%), Positives = 55/78 (70%), Gaps = 0/78 (0%)
Query 16 LRPAVRCPTQKYNIKVREGRGFTLEELKAVGVSPRVALSIGIAVDHRRRNRSQESLNNNI 75
+RP V T YN+KVR G+GFTLEELKA G+ ++A +IGI+VDH R+NRS E N+
Sbjct 53 IRPVVHAQTLTYNMKVRAGKGFTLEELKAAGIPKKLAPTIGISVDHHRKNRSLEGFQTNV 112
Query 76 NRLKLYLNKLVLFQRGSK 93
RLK Y KLV+F R ++
Sbjct 113 QRLKTYKAKLVIFPRCAR 130
> ECU03g0320
Length=163
Score = 80.5 bits (197), Expect = 7e-16, Method: Compositional matrix adjust.
Identities = 38/78 (48%), Positives = 52/78 (66%), Gaps = 0/78 (0%)
Query 12 PVDLLRPAVRCPTQKYNIKVREGRGFTLEELKAVGVSPRVALSIGIAVDHRRRNRSQESL 71
P+ LRP VRCPT KYN R GRGFT E + G+ R A +GIAVD RRR+ +QE+
Sbjct 47 PLKKLRPIVRCPTIKYNRNERLGRGFTAAECEKAGLDYRHARRLGIAVDLRRRDTNQEAF 106
Query 72 NNNINRLKLYLNKLVLFQ 89
+ N+ R+K YL K+ +++
Sbjct 107 DKNVERIKTYLGKITIYE 124
> Hs14784147
Length=144
Score = 73.6 bits (179), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 37/81 (45%), Positives = 53/81 (65%), Gaps = 1/81 (1%)
Query 18 PAVRCPTQKYNIKVREGRGFTLEELKAVGVSPRVALSIGIAVDHRRRNRSQESLNNNINR 77
P VRCP +++ K + GRG +LEEL+ G+ +VA +IGI+ D RRRN+S ++L + R
Sbjct 31 PIVRCPMLRHHYKAQAGRGLSLEELRVAGIYKKVAQTIGISEDARRRNQSTQALQAKVQR 90
Query 78 LKLYLNKLVLFQRGS-KPKKG 97
LK + L+LF R PKKG
Sbjct 91 LKEDRSSLILFPRKPLAPKKG 111
> Hs22045017
Length=180
Score = 63.9 bits (154), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 34/64 (53%), Positives = 43/64 (67%), Gaps = 1/64 (1%)
Query 39 LEELKAVGVSPRVALSIGIAVDHRRRNRSQESLNNNINRLKLYLNKLVLFQRGS-KPKKG 97
LEELK G+ VA + GI++D RRRN+S ESL N+ RLK Y +KL+LF R PKKG
Sbjct 43 LEELKVAGIHKNVAQTTGISIDWRRRNKSTESLQGNVQRLKEYNSKLILFPRKPLAPKKG 102
Query 98 LAGV 101
+ V
Sbjct 103 DSSV 106
> Hs17466264
Length=161
Score = 37.4 bits (85), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 23/32 (71%), Gaps = 1/32 (3%)
Query 17 RPA-VRCPTQKYNIKVREGRGFTLEELKAVGV 47
PA +RC +Y+ KV G+GF++EEL+ VG+
Sbjct 32 HPAHIRCCMVRYHTKVHAGQGFSMEELRVVGI 63
> 7296399
Length=446
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 24/79 (30%), Positives = 37/79 (46%), Gaps = 10/79 (12%)
Query 29 IKVREGRGFTLEELKAVGVSPRVA---LSIGIAVDHRRRNRSQESLNNNINRLKLYLNKL 85
+ V E R F ++ KAV V A + +A DHR ++ +NRL++Y+N L
Sbjct 94 VAVAESRRFMIDCFKAVKVPQAHAEAQADLLVAADHRGH------FSHGMNRLEMYINDL 147
Query 86 VLFQR-GSKPKKGLAGVPA 103
+ G+ K L PA
Sbjct 148 AINSTDGAAVPKILKETPA 166
> At4g26450
Length=1248
Score = 27.3 bits (59), Expect = 7.6, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 60 DHRRRNRSQESLNNNINRLKLYLNKLVLFQR 90
+ R N + NNNI+ +KL L KLV F +
Sbjct 1215 EERDTNSAIRKKNNNISSMKLLLKKLVTFSK 1245
> At4g01440
Length=365
Score = 26.9 bits (58), Expect = 9.2, Method: Composition-based stats.
Identities = 12/50 (24%), Positives = 26/50 (52%), Gaps = 0/50 (0%)
Query 25 QKYNIKVREGRGFTLEELKAVGVSPRVALSIGIAVDHRRRNRSQESLNNN 74
+K N+K + G G + L +G + + + G+ + R+ + + +NNN
Sbjct 124 EKLNMKSKAGMGMVMGALICIGGALLLTMYKGVPLTKLRKLETHQLINNN 173
Lambda K H
0.318 0.136 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1170944580
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40