bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
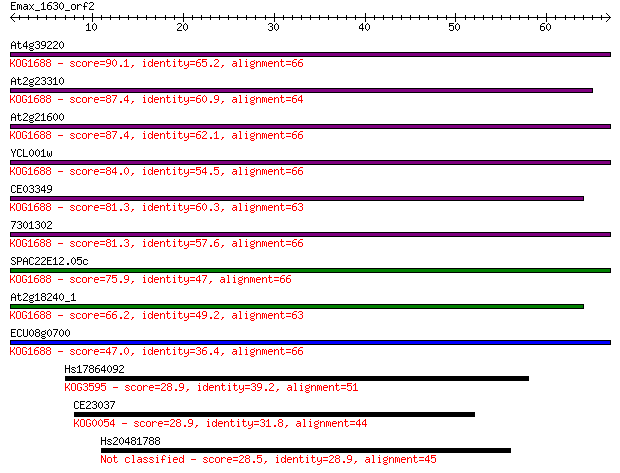
Query= Emax_1630_orf2
Length=66
Score E
Sequences producing significant alignments: (Bits) Value
At4g39220 90.1 9e-19
At2g23310 87.4 5e-18
At2g21600 87.4 6e-18
YCL001w 84.0 6e-17
CE03349 81.3 4e-16
7301302 81.3 5e-16
SPAC22E12.05c 75.9 2e-14
At2g18240_1 66.2 1e-11
ECU08g0700 47.0 9e-06
Hs17864092 28.9 2.2
CE23037 28.9 2.7
Hs20481788 28.5 3.1
> At4g39220
Length=191
Score = 90.1 bits (222), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 43/66 (65%), Positives = 53/66 (80%), Gaps = 0/66 (0%)
Query 1 FKAWVSGMRALLIAISMTFFQVFDLPVFWPILLIYFILLFILTMKQQVKKMIKYKYLPFS 60
FK W S +A IA MTFF VFD+PVFWPILL Y+I+LF+LTM++Q+ MIKYKY+PFS
Sbjct 115 FKFWYSMTKAFCIAFLMTFFSVFDVPVFWPILLCYWIVLFVLTMRRQIAHMIKYKYIPFS 174
Query 61 WGKQTY 66
+GKQ Y
Sbjct 175 FGKQKY 180
> At2g23310
Length=211
Score = 87.4 bits (215), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 39/64 (60%), Positives = 55/64 (85%), Gaps = 0/64 (0%)
Query 1 FKAWVSGMRALLIAISMTFFQVFDLPVFWPILLIYFILLFILTMKQQVKKMIKYKYLPFS 60
FK W+S +RA +I MTFF+VFD+PVFWPILL Y+++LF LTM++Q++ MIKY+Y+PFS
Sbjct 136 FKFWLSIIRAFIIGFMMTFFEVFDVPVFWPILLFYWVMLFFLTMRKQIQHMIKYRYVPFS 195
Query 61 WGKQ 64
+GK+
Sbjct 196 FGKK 199
> At2g21600
Length=195
Score = 87.4 bits (215), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 41/66 (62%), Positives = 52/66 (78%), Gaps = 0/66 (0%)
Query 1 FKAWVSGMRALLIAISMTFFQVFDLPVFWPILLIYFILLFILTMKQQVKKMIKYKYLPFS 60
FK W S +A IA MTFF VFD+PVFWPILL Y+++LF+LTM++Q+ MIK+KY+PFS
Sbjct 114 FKFWYSMTKAFCIAFLMTFFSVFDVPVFWPILLCYWVVLFVLTMRRQIAHMIKHKYIPFS 173
Query 61 WGKQTY 66
GKQ Y
Sbjct 174 IGKQKY 179
> YCL001w
Length=188
Score = 84.0 bits (206), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 36/66 (54%), Positives = 53/66 (80%), Gaps = 0/66 (0%)
Query 1 FKAWVSGMRALLIAISMTFFQVFDLPVFWPILLIYFILLFILTMKQQVKKMIKYKYLPFS 60
FK W + +RA +I++ ++ F +FD+PVFWPILL+YFILLF LTM++Q++ MIKY+Y+P
Sbjct 118 FKFWYNSIRATVISLLLSLFSIFDIPVFWPILLMYFILLFFLTMRRQIQHMIKYRYIPLD 177
Query 61 WGKQTY 66
GK+ Y
Sbjct 178 IGKKKY 183
> CE03349
Length=191
Score = 81.3 bits (199), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 38/63 (60%), Positives = 51/63 (80%), Gaps = 0/63 (0%)
Query 1 FKAWVSGMRALLIAISMTFFQVFDLPVFWPILLIYFILLFILTMKQQVKKMIKYKYLPFS 60
FK W S M+A LIAI+ TFF+ FD+PVFWPIL++YF +L LT+K+Q+ MIKY+Y+PF+
Sbjct 114 FKFWHSFMKATLIAITCTFFEFFDVPVFWPILVMYFFILTFLTLKRQIMHMIKYRYIPFT 173
Query 61 WGK 63
GK
Sbjct 174 VGK 176
> 7301302
Length=203
Score = 81.3 bits (199), Expect = 5e-16, Method: Compositional matrix adjust.
Identities = 38/66 (57%), Positives = 50/66 (75%), Gaps = 0/66 (0%)
Query 1 FKAWVSGMRALLIAISMTFFQVFDLPVFWPILLIYFILLFILTMKQQVKKMIKYKYLPFS 60
FK W+S ++ LI + TFF F++PVFWPIL++YFI LF +TMK+Q+K MIKYKYLPF+
Sbjct 119 FKFWLSVAKSTLIGLICTFFDFFNVPVFWPILVMYFITLFCITMKRQIKHMIKYKYLPFT 178
Query 61 WGKQTY 66
K Y
Sbjct 179 RNKPRY 184
> SPAC22E12.05c
Length=184
Score = 75.9 bits (185), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 31/66 (46%), Positives = 50/66 (75%), Gaps = 0/66 (0%)
Query 1 FKAWVSGMRALLIAISMTFFQVFDLPVFWPILLIYFILLFILTMKQQVKKMIKYKYLPFS 60
FK W S MRA L A+ +FF++FD+PVFWPIL++Y+++L ++Q++ M+KY+Y+PF
Sbjct 116 FKFWYSSMRATLFALVASFFRIFDVPVFWPILVVYYLVLSFFCFRRQIQHMLKYRYVPFD 175
Query 61 WGKQTY 66
GK+ +
Sbjct 176 IGKKKF 181
> At2g18240_1
Length=180
Score = 66.2 bits (160), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 31/63 (49%), Positives = 43/63 (68%), Gaps = 0/63 (0%)
Query 1 FKAWVSGMRALLIAISMTFFQVFDLPVFWPILLIYFILLFILTMKQQVKKMIKYKYLPFS 60
FK W + +A ++A MTFF D+PVFWPILL Y+++L+ LTMK+ + M KY+Y PF
Sbjct 118 FKFWYAATKAFVVAFVMTFFSFLDVPVFWPILLCYWLVLYSLTMKRLIVHMFKYRYFPFD 177
Query 61 WGK 63
K
Sbjct 178 VRK 180
> ECU08g0700
Length=166
Score = 47.0 bits (110), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 24/66 (36%), Positives = 39/66 (59%), Gaps = 0/66 (0%)
Query 1 FKAWVSGMRALLIAISMTFFQVFDLPVFWPILLIYFILLFILTMKQQVKKMIKYKYLPFS 60
F W+ + + +A+ T+F + D+PV+ PIL++YFI + T K+ + M KY Y PF
Sbjct 99 FDFWMFVTKIVGMALIGTYFGILDVPVYTPILVVYFIFMVGYTAKRLIAHMKKYNYNPFL 158
Query 61 WGKQTY 66
K+ Y
Sbjct 159 QSKEYY 164
> Hs17864092
Length=4024
Score = 28.9 bits (63), Expect = 2.2, Method: Composition-based stats.
Identities = 20/54 (37%), Positives = 28/54 (51%), Gaps = 3/54 (5%)
Query 7 GMRALLIAISMTFFQVFDLPVFWPIL---LIYFILLFILTMKQQVKKMIKYKYL 57
G R + I S+ FF + DL P+ L +FI LFIL+++ K I K L
Sbjct 3202 GYRPIAIHSSILFFSLADLANIEPMYQYSLTWFINLFILSIENSEKSEILAKRL 3255
> CE23037
Length=1144
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 14/44 (31%), Positives = 27/44 (61%), Gaps = 3/44 (6%)
Query 8 MRALLIAISMTFFQVFDLPVFWPILLIYFILLFILTMKQQVKKM 51
M +LI+IS F V P+ IL+ YF++++ + +Q+K++
Sbjct 711 MILVLISISTPIFLVCAAPL---ILIYYFVMIYYIPTSRQLKRL 751
> Hs20481788
Length=543
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 13/45 (28%), Positives = 23/45 (51%), Gaps = 0/45 (0%)
Query 11 LLIAISMTFFQVFDLPVFWPILLIYFILLFILTMKQQVKKMIKYK 55
+L + S F+ +F LP +P+LLI+ + Q+ K Y+
Sbjct 219 MLCSFSHLFYPLFHLPTLFPLLLIFLANVTFNYSNQKPKPSTSYQ 263
Lambda K H
0.338 0.147 0.489
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1163608362
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40