bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1653_orf1
Length=68
Score E
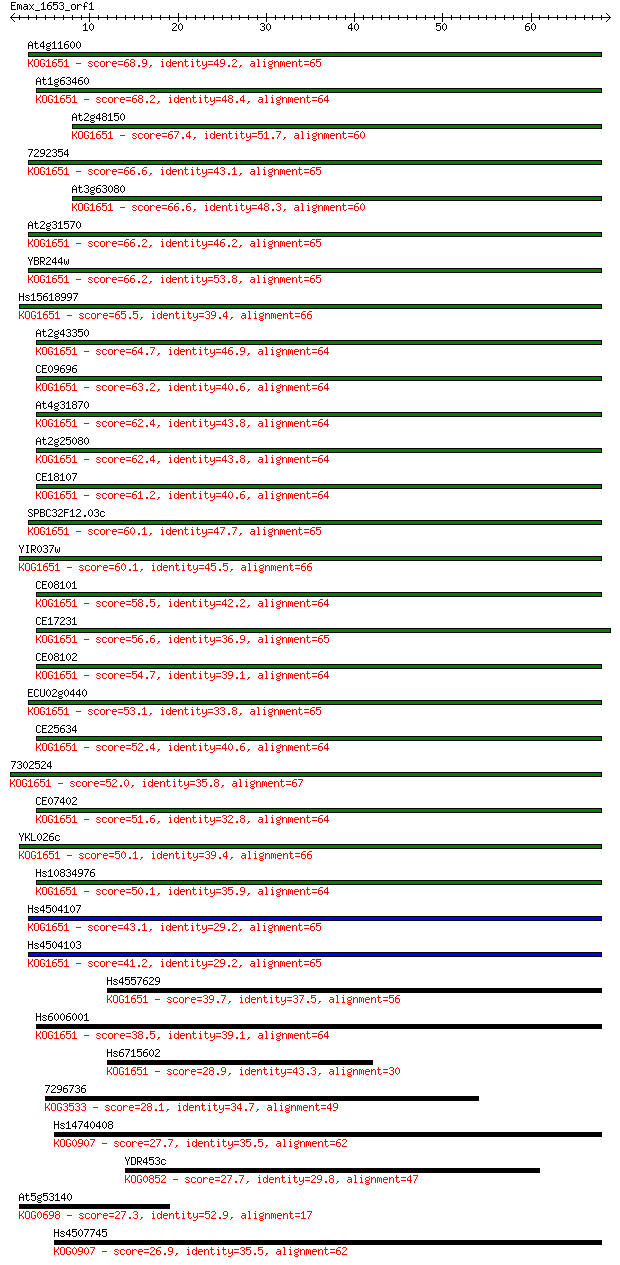
Sequences producing significant alignments: (Bits) Value
At4g11600 68.9 2e-12
At1g63460 68.2 3e-12
At2g48150 67.4 7e-12
7292354 66.6 1e-11
At3g63080 66.6 1e-11
At2g31570 66.2 1e-11
YBR244w 66.2 1e-11
Hs15618997 65.5 2e-11
At2g43350 64.7 4e-11
CE09696 63.2 1e-10
At4g31870 62.4 2e-10
At2g25080 62.4 2e-10
CE18107 61.2 4e-10
SPBC32F12.03c 60.1 1e-09
YIR037w 60.1 1e-09
CE08101 58.5 3e-09
CE17231 56.6 1e-08
CE08102 54.7 4e-08
ECU02g0440 53.1 1e-07
CE25634 52.4 2e-07
7302524 52.0 3e-07
CE07402 51.6 4e-07
YKL026c 50.1 1e-06
Hs10834976 50.1 1e-06
Hs4504107 43.1 1e-04
Hs4504103 41.2 5e-04
Hs4557629 39.7 0.001
Hs6006001 38.5 0.003
Hs6715602 28.9 2.3
7296736 28.1 3.8
Hs14740408 27.7 5.2
YDR453c 27.7 5.3
At5g53140 27.3 7.1
Hs4507745 26.9 9.1
> At4g11600
Length=169
Score = 68.9 bits (167), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 32/65 (49%), Positives = 40/65 (61%), Gaps = 0/65 (0%)
Query 3 FYSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAF 62
Y K AKG ++ + YKGKVLLI N A++CG T + E ++ EKY GFEILAF
Sbjct 10 LYDFTVKDAKGNDVDLSIYKGKVLLIVNVASQCGLTNSNYTELAQLYEKYKGHGFEILAF 69
Query 63 PTLQF 67
P QF
Sbjct 70 PCNQF 74
> At1g63460
Length=167
Score = 68.2 bits (165), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 31/64 (48%), Positives = 40/64 (62%), Gaps = 0/64 (0%)
Query 4 YSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAFP 63
Y L + AKG + + YK KVLLI N A+KCG T + E +E+ +Y D+G EILAFP
Sbjct 10 YELSIEDAKGNNLALSQYKDKVLLIVNVASKCGMTNSNYTELNELYNRYKDKGLEILAFP 69
Query 64 TLQF 67
QF
Sbjct 70 CNQF 73
> At2g48150
Length=171
Score = 67.4 bits (163), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 31/60 (51%), Positives = 41/60 (68%), Gaps = 0/60 (0%)
Query 8 AKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAFPTLQF 67
K + G+++ M Y+GKVLLI N A+KCGFT+ + + E+ KY DQ FEILAFP QF
Sbjct 17 VKDSSGKDLNMSIYQGKVLLIVNVASKCGFTETNYTQLTELYRKYKDQDFEILAFPCNQF 76
> 7292354
Length=169
Score = 66.6 bits (161), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 28/65 (43%), Positives = 42/65 (64%), Gaps = 0/65 (0%)
Query 3 FYSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAF 62
Y K G ++ ++ YKGKV+L+ N A+KCG TK + ++ ++KEKYG++G IL F
Sbjct 13 IYEFTVKDTHGNDVSLEKYKGKVVLVVNIASKCGLTKNNYEKLTDLKEKYGERGLVILNF 72
Query 63 PTLQF 67
P QF
Sbjct 73 PCNQF 77
> At3g63080
Length=173
Score = 66.6 bits (161), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 29/60 (48%), Positives = 41/60 (68%), Gaps = 0/60 (0%)
Query 8 AKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAFPTLQF 67
K + G+E+ + Y+GKVLL+ N A+KCGFT+ + + E+ KY DQGF +LAFP QF
Sbjct 19 VKDSSGKEVDLSVYQGKVLLVVNVASKCGFTESNYTQLTELYRKYKDQGFVVLAFPCNQF 78
> At2g31570
Length=169
Score = 66.2 bits (160), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 30/65 (46%), Positives = 39/65 (60%), Gaps = 0/65 (0%)
Query 3 FYSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAF 62
Y K G ++ + YKGK LL+ N A+KCG T + KE + + EKY +QG EILAF
Sbjct 9 IYDFTVKDIGGNDVSLDQYKGKTLLVVNVASKCGLTDANYKELNVLYEKYKEQGLEILAF 68
Query 63 PTLQF 67
P QF
Sbjct 69 PCNQF 73
> YBR244w
Length=162
Score = 66.2 bits (160), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 35/65 (53%), Positives = 42/65 (64%), Gaps = 1/65 (1%)
Query 3 FYSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAF 62
FY L+ K KGE KGKV+LI N A+KCGFT Q+ KE E+ +KY D+GF IL F
Sbjct 5 FYDLECKDKKGESFKFDQLKGKVVLIVNVASKCGFTPQY-KELEELYKKYQDKGFVILGF 63
Query 63 PTLQF 67
P QF
Sbjct 64 PCNQF 68
> Hs15618997
Length=187
Score = 65.5 bits (158), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 40/66 (60%), Gaps = 0/66 (0%)
Query 2 DFYSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILA 61
DFY KA +G+ + ++ Y+G V L+ N A++CGFT QH + +++ G F +LA
Sbjct 24 DFYDFKAVNIRGKLVSLEKYRGSVSLVVNVASECGFTDQHYRALQQLQRDLGPHHFNVLA 83
Query 62 FPTLQF 67
FP QF
Sbjct 84 FPCNQF 89
> At2g43350
Length=206
Score = 64.7 bits (156), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 30/64 (46%), Positives = 42/64 (65%), Gaps = 0/64 (0%)
Query 4 YSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAFP 63
Y++ K +G+++ + + GKVLLI N A+KCG T + KE + + KY QGFEILAFP
Sbjct 49 YNISVKDIEGKDVSLSKFTGKVLLIVNVASKCGLTHGNYKEMNILYAKYKTQGFEILAFP 108
Query 64 TLQF 67
QF
Sbjct 109 CNQF 112
> CE09696
Length=163
Score = 63.2 bits (152), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 39/64 (60%), Gaps = 0/64 (0%)
Query 4 YSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAFP 63
Y K A G+++ + YKGKVL+I N A++CG T ++ + E+ + Y G E+LAFP
Sbjct 5 YDFNVKNANGDDVSLSDYKGKVLIIVNVASQCGLTNKNYTQLKELLDVYKKDGLEVLAFP 64
Query 64 TLQF 67
QF
Sbjct 65 CNQF 68
> At4g31870
Length=230
Score = 62.4 bits (150), Expect = 2e-10, Method: Composition-based stats.
Identities = 28/64 (43%), Positives = 39/64 (60%), Gaps = 0/64 (0%)
Query 4 YSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAFP 63
+ K G ++ + +KGK LLI N A++CG T + E ++ EKY +QGFEILAFP
Sbjct 77 HDFTVKDIDGNDVSLDKFKGKPLLIVNVASRCGLTSSNYSELSQLYEKYKNQGFEILAFP 136
Query 64 TLQF 67
QF
Sbjct 137 CNQF 140
> At2g25080
Length=236
Score = 62.4 bits (150), Expect = 2e-10, Method: Composition-based stats.
Identities = 28/64 (43%), Positives = 39/64 (60%), Gaps = 0/64 (0%)
Query 4 YSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAFP 63
+ K G+++ + +KGKV+LI N A++CG T + E + EKY QGFEILAFP
Sbjct 80 HDFTVKDIDGKDVALNKFKGKVMLIVNVASRCGLTSSNYSELSHLYEKYKTQGFEILAFP 139
Query 64 TLQF 67
QF
Sbjct 140 CNQF 143
> CE18107
Length=163
Score = 61.2 bits (147), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 40/64 (62%), Gaps = 0/64 (0%)
Query 4 YSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAFP 63
+ + K A+GE+ + Y+GKVL+I N A++CG T + +F E+ + Y G E+LAFP
Sbjct 5 HGITVKNAQGEDTPLSNYQGKVLIIVNVASQCGLTNSNYNQFKELLDVYKKDGLEVLAFP 64
Query 64 TLQF 67
QF
Sbjct 65 CNQF 68
> SPBC32F12.03c
Length=158
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 31/65 (47%), Positives = 38/65 (58%), Gaps = 1/65 (1%)
Query 3 FYSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAF 62
FY L K G KGKV+L+ NTA+KCGFT Q+ K + +KY D+GF IL F
Sbjct 4 FYDLAPKDKDGNPFPFSNLKGKVVLVVNTASKCGFTPQY-KGLEALYQKYKDRGFIILGF 62
Query 63 PTLQF 67
P QF
Sbjct 63 PCNQF 67
> YIR037w
Length=163
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 30/66 (45%), Positives = 40/66 (60%), Gaps = 1/66 (1%)
Query 2 DFYSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILA 61
+FY L KG+ KGKV+LI N A+KCGFT Q+ KE + ++Y D+GF I+
Sbjct 3 EFYKLAPVDKKGQPFPFDQLKGKVVLIVNVASKCGFTPQY-KELEALYKRYKDEGFTIIG 61
Query 62 FPTLQF 67
FP QF
Sbjct 62 FPCNQF 67
> CE08101
Length=223
Score = 58.5 bits (140), Expect = 3e-09, Method: Composition-based stats.
Identities = 27/64 (42%), Positives = 42/64 (65%), Gaps = 1/64 (1%)
Query 4 YSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAFP 63
Y + +T +GE + Y+G+VLL+ N AT C +T+Q+ +F+ + EKY QGF ++AFP
Sbjct 41 YDFQVETLQGEYTDLSQYRGQVLLMVNVATFCAYTQQY-TDFNPLIEKYQSQGFTLIAFP 99
Query 64 TLQF 67
QF
Sbjct 100 CNQF 103
> CE17231
Length=188
Score = 56.6 bits (135), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 39/65 (60%), Gaps = 0/65 (0%)
Query 4 YSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAFP 63
Y +AK G+ + M+ Y+ KV+L +N A+ CG+T + F E+ Y ++GF + AFP
Sbjct 32 YQFQAKNIDGKMVSMEKYRDKVVLFTNVASYCGYTDSNYNAFKELDGIYREKGFRVAAFP 91
Query 64 TLQFK 68
QF+
Sbjct 92 CNQFE 96
> CE08102
Length=224
Score = 54.7 bits (130), Expect = 4e-08, Method: Composition-based stats.
Identities = 25/64 (39%), Positives = 41/64 (64%), Gaps = 1/64 (1%)
Query 4 YSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAFP 63
+ + +T +GE + Y+GKV+L+ N AT C +T+Q+ +F+ + EKY QG ++AFP
Sbjct 42 FDFQIETLQGEYTDLSQYRGKVILLVNVATFCAYTQQY-TDFNPMLEKYQAQGLTLVAFP 100
Query 64 TLQF 67
QF
Sbjct 101 CNQF 104
> ECU02g0440
Length=177
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 39/65 (60%), Gaps = 0/65 (0%)
Query 3 FYSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAF 62
FY L A+ G E+ + +++G V++I+N A+ C F + + K F + +K+ +G IL F
Sbjct 10 FYGLSARGWDGSEVSLGSFRGCVIMIANVASSCKFAESNYKSFAGLLDKFYRKGLRILLF 69
Query 63 PTLQF 67
P Q+
Sbjct 70 PCNQY 74
> CE25634
Length=199
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/64 (40%), Positives = 39/64 (60%), Gaps = 1/64 (1%)
Query 4 YSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAFP 63
+ + +T KG+ + Y+GKV L+ N AT C +T+Q+ +F+ I +KY QG I AFP
Sbjct 41 FDFQIETLKGDYTDLSQYRGKVTLLVNVATFCAYTQQY-TDFNPILDKYQKQGLVIAAFP 99
Query 64 TLQF 67
QF
Sbjct 100 CNQF 103
> 7302524
Length=160
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 34/67 (50%), Gaps = 0/67 (0%)
Query 1 MDFYSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEIL 60
+ ++L + G + + + G VLLI N A+KCG T + E+Y DQG IL
Sbjct 5 LTIHALTVRDTFGNPVQLDTFAGHVLLIVNIASKCGLTLSQYNGLRYLLEEYEDQGLRIL 64
Query 61 AFPTLQF 67
FP QF
Sbjct 65 NFPCNQF 71
> CE07402
Length=193
Score = 51.6 bits (122), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 33/64 (51%), Gaps = 0/64 (0%)
Query 4 YSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAFP 63
Y + G+ + + Y G V++I N A+ CG T + KE + +KY +G + AFP
Sbjct 34 YDFSVRDNSGDLVSLDKYSGLVVIIVNVASYCGLTNSNYKELKSLNDKYHLRGLRVAAFP 93
Query 64 TLQF 67
QF
Sbjct 94 CNQF 97
> YKL026c
Length=167
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 26/66 (39%), Positives = 34/66 (51%), Gaps = 1/66 (1%)
Query 2 DFYSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILA 61
+FYS G + + KV+LI N A+ C FT Q+ KE + EKY G I+A
Sbjct 3 EFYSFSPIDENGNPFPFNSLRNKVVLIVNVASHCAFTPQY-KELEYLYEKYKSHGLVIVA 61
Query 62 FPTLQF 67
FP QF
Sbjct 62 FPCGQF 67
> Hs10834976
Length=201
Score = 50.1 bits (118), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 38/65 (58%), Gaps = 1/65 (1%)
Query 4 YSLKAKT-AKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAF 62
Y+ A+ A GE + + + +GKVLLI N A+ G T + + +E++ + G +G +L F
Sbjct 15 YAFSARPLAGGEPVSLGSLRGKVLLIENVASLXGTTVRDYTQMNELQRRLGPRGLVVLGF 74
Query 63 PTLQF 67
P QF
Sbjct 75 PCNQF 79
> Hs4504107
Length=197
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 35/65 (53%), Gaps = 0/65 (0%)
Query 3 FYSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAF 62
+ AK G + + Y+G V +++N A++ G T+ + + ++ +Y + G ILAF
Sbjct 41 MHEFSAKDIDGHMVNLDKYRGFVCIVTNVASQXGKTEVNYTQLVDLHARYAECGLRILAF 100
Query 63 PTLQF 67
P QF
Sbjct 101 PCNQF 105
> Hs4504103
Length=190
Score = 41.2 bits (95), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 36/65 (55%), Gaps = 1/65 (1%)
Query 3 FYSLKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAF 62
FY L A + GE++ ++G+ +LI N A+ G T + + +E++ ++ + +L F
Sbjct 8 FYDLSAISLDGEKVDFNTFRGRAVLIENVASLXGTTTRDFTQLNELQCRFPRR-LVVLGF 66
Query 63 PTLQF 67
P QF
Sbjct 67 PCNQF 71
> Hs4557629
Length=221
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 21/56 (37%), Positives = 30/56 (53%), Gaps = 1/56 (1%)
Query 12 KGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAFPTLQF 67
K E + + Y GK +L N AT CG T Q+ E + ++E+ G +L FP QF
Sbjct 50 KNEYVSFKQYVGKHILFVNVATYCGLTAQY-PELNALQEELKPYGLVVLGFPCNQF 104
> Hs6006001
Length=226
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 25/65 (38%), Positives = 34/65 (52%), Gaps = 2/65 (3%)
Query 4 YSLKAKTAKGEE-ICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGFEILAF 62
Y A T GEE I + Y GK +L N A+ G T Q++ E + ++E+ G IL F
Sbjct 41 YEYGALTIDGEEYIPFKQYAGKYVLFVNVASYUGLTGQYI-ELNALQEELAPFGLVILGF 99
Query 63 PTLQF 67
P QF
Sbjct 100 PCNQF 104
> Hs6715602
Length=100
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 12 KGEEICMQAYKGKVLLISNTATKCGFTKQH 41
K E + + Y GK +L N AT CG T Q+
Sbjct 50 KNEYVSFKQYVGKHILFVNVATYCGLTAQY 79
> 7296736
Length=2837
Score = 28.1 bits (61), Expect = 3.8, Method: Composition-based stats.
Identities = 17/53 (32%), Positives = 30/53 (56%), Gaps = 4/53 (7%)
Query 5 SLKAKTAKGEEICMQAYKGKVL----LISNTATKCGFTKQHLKEFHEIKEKYG 53
SL A A GE+I +++ + ++L LI N +T+ K H+ E + K++ G
Sbjct 2772 SLAAVDADGEQIELRSMQAQLLDTQLLIKNLSTQLHELKDHMTEQRKQKQRLG 2824
> Hs14740408
Length=105
Score = 27.7 bits (60), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 22/77 (28%), Positives = 34/77 (44%), Gaps = 17/77 (22%)
Query 6 LKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGF-------- 57
+++KTA E + A K++++ +AT CG K FH + EKY + F
Sbjct 5 IESKTAFQEA--LDAAGDKLVVVDFSATWCGPCKMIKPFFHSLSEKYSNVIFLEVDVDDC 62
Query 58 -------EILAFPTLQF 67
E+ PT QF
Sbjct 63 QDVASECEVKCMPTFQF 79
> YDR453c
Length=196
Score = 27.7 bits (60), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 14/48 (29%), Positives = 26/48 (54%), Gaps = 1/48 (2%)
Query 14 EEICMQAYKGKVLLISNTATKCGFT-KQHLKEFHEIKEKYGDQGFEIL 60
EEI ++ YKGK ++++ F + F + +K+ DQG ++L
Sbjct 23 EEISLEKYKGKYVVLAFVPLAFSFVCPTEIVAFSDAAKKFEDQGAQVL 70
> At5g53140
Length=307
Score = 27.3 bits (59), Expect = 7.1, Method: Composition-based stats.
Identities = 9/17 (52%), Positives = 13/17 (76%), Gaps = 0/17 (0%)
Query 2 DFYSLKAKTAKGEEICM 18
DFY +KA T +G+ +CM
Sbjct 3 DFYDIKASTIEGQAVCM 19
> Hs4507745
Length=105
Score = 26.9 bits (58), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 22/77 (28%), Positives = 34/77 (44%), Gaps = 17/77 (22%)
Query 6 LKAKTAKGEEICMQAYKGKVLLISNTATKCGFTKQHLKEFHEIKEKYGDQGF-------- 57
+++KTA E + A K++++ +AT CG K FH + EKY + F
Sbjct 5 IESKTAFQEA--LDAAGDKLVVVDFSATWCGPCKMINPFFHSLSEKYSNVIFLEVDVDDC 62
Query 58 -------EILAFPTLQF 67
E+ PT QF
Sbjct 63 QDVASECEVKCTPTFQF 79
Lambda K H
0.321 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1203543208
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40