bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1721_orf1
Length=129
Score E
Sequences producing significant alignments: (Bits) Value
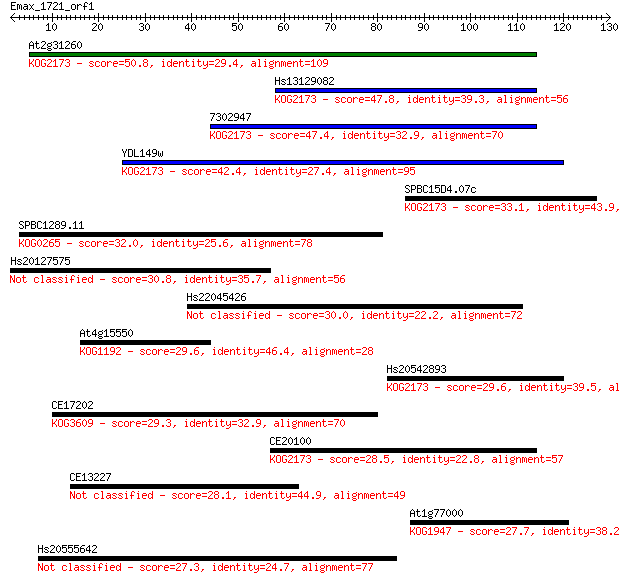
At2g31260 50.8 7e-07
Hs13129082 47.8 5e-06
7302947 47.4 8e-06
YDL149w 42.4 2e-04
SPBC15D4.07c 33.1 0.14
SPBC1289.11 32.0 0.27
Hs20127575 30.8 0.58
Hs22045426 30.0 1.2
At4g15550 29.6 1.3
Hs20542893 29.6 1.6
CE17202 29.3 2.0
CE20100 28.5 2.9
CE13227 28.1 3.9
At1g77000 27.7 5.4
Hs20555642 27.3 6.6
> At2g31260
Length=865
Score = 50.8 bits (120), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 32/109 (29%), Positives = 55/109 (50%), Gaps = 2/109 (1%)
Query 5 PFSPWGPWQTLCTVYVLSLSWVVLFSLFSVFNKCKDAVALRLYFRSRLYIHCDTALQLMG 64
P SP+ + Y+ S LF F + KD + R ++ + L++ D + M
Sbjct 147 PLSPFTLTTAIIVGYLALFSVYWLFCFLRFFAQLKDTLDFRHFYYNNLHV-TDNEILTMP 205
Query 65 WPEVAALLLRAQQQHPFCLVKQNLSVLDLVSVIMRRDNYIIALTNKEVL 113
W V +++ Q C+VK +LS D+V +MR++NY+I + NK +L
Sbjct 206 WATVLEKVVQLQSSQCLCVVK-DLSAHDMVMRLMRKENYLIGMLNKGLL 253
> Hs13129082
Length=446
Score = 47.8 bits (112), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 36/56 (64%), Gaps = 0/56 (0%)
Query 58 TALQLMGWPEVAALLLRAQQQHPFCLVKQNLSVLDLVSVIMRRDNYIIALTNKEVL 113
+AL W EV A +++ Q++H C+ K+ L+ LD+ I+R NY++AL NK +L
Sbjct 2 SALPYCTWQEVQARIVQTQKEHQICIHKRELTELDIYHRILRFQNYMVALVNKSLL 57
> 7302947
Length=845
Score = 47.4 bits (111), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 23/70 (32%), Positives = 42/70 (60%), Gaps = 1/70 (1%)
Query 44 LRLYFRSRLYIHCDTALQLMGWPEVAALLLRAQQQHPFCLVKQNLSVLDLVSVIMRRDNY 103
++ ++ S L+I D+ L W EV + R Q + C+ K++L+ LD+ ++R NY
Sbjct 205 IKRFYNSALHIE-DSDLDNFTWHEVQQRIRRVQAEQHMCIDKESLTELDIYHRVLRFKNY 263
Query 104 IIALTNKEVL 113
++AL NK++L
Sbjct 264 LVALMNKQLL 273
> YDL149w
Length=997
Score = 42.4 bits (98), Expect = 2e-04, Method: Composition-based stats.
Identities = 26/101 (25%), Positives = 49/101 (48%), Gaps = 7/101 (6%)
Query 25 WVVLFSLFSVFNKCKDAVALRLYFRSRLYIHCDTALQLMGWPEVAALLLRAQQQHPFCL- 83
+ V+ + ++ + L+ +++ L I D LQ + W V L+ + Q+
Sbjct 384 FFVILKIVQLYFDVQKLSELQNFYKYLLNIS-DDELQTLPWQNVIQQLMYLKDQNAMTAN 442
Query 84 -----VKQNLSVLDLVSVIMRRDNYIIALTNKEVLGRALPL 119
K + D+ + IMRR+NY+IAL N ++L +LP+
Sbjct 443 VVEVKAKNRIDAHDVANRIMRRENYLIALYNSDILNLSLPI 483
> SPBC15D4.07c
Length=702
Score = 33.1 bits (74), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 27/44 (61%), Gaps = 4/44 (9%)
Query 86 QNLSVLDLVSV---IMRRDNYIIALTNKEVLGRALPLYVHPKLL 126
N+ LD ++ IMR+DNY IAL N ++ LPL +H ++L
Sbjct 329 HNMKRLDAYAIANRIMRKDNYFIALINNGIINIELPL-LHRRIL 371
> SPBC1289.11
Length=340
Score = 32.0 bits (71), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 20/81 (24%), Positives = 34/81 (41%), Gaps = 3/81 (3%)
Query 3 VSPFSPWGPWQTLCTVYVLSLSWVVLFSL--FSVFNKCKDAVALRLYFRSRLYIHCDTA- 59
V+ F P G + + L W V + + V N CK A+ + R ++C ++
Sbjct 55 VARFDPSGSYFASGGMDRQILLWNVFGDVKNYGVLNGCKGAITDLQWSRDSRVVYCSSSD 114
Query 60 LQLMGWPEVAALLLRAQQQHP 80
LM W V+ +R + H
Sbjct 115 THLMSWDAVSGQKIRKHKGHA 135
> Hs20127575
Length=285
Score = 30.8 bits (68), Expect = 0.58, Method: Compositional matrix adjust.
Identities = 20/63 (31%), Positives = 31/63 (49%), Gaps = 7/63 (11%)
Query 1 LRVSPFSPWG---PWQTLCTVYVLS-LSWVVLFSLFSVFNKC---KDAVALRLYFRSRLY 53
+R F PW +Q LC+ L+ L WV F+ F ++ KC D LR Y++ +
Sbjct 217 IRFHSFYPWHTGRDYQQLCSQQDLAMLPWVREFNKFDLYTKCPDLPDVDKLRPYYQGLID 276
Query 54 IHC 56
+C
Sbjct 277 KYC 279
> Hs22045426
Length=360
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 16/74 (21%), Positives = 34/74 (45%), Gaps = 18/74 (24%)
Query 39 KDAVALRLYFRSRLYIHCDTALQLMGWPEVAALLLRAQQQHPFCLVKQNLSVLDLVSVI- 97
KD + +R Y +Y+H ++ +++H +C + + + LD S I
Sbjct 27 KDVLRVRFYHEKCIYVHKES----------------TKERHGYCTLGEAFNRLDFSSAIQ 70
Query 98 -MRRDNYIIALTNK 110
+RR NY++ ++
Sbjct 71 DIRRFNYVVKAASE 84
> At4g15550
Length=458
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 20/32 (62%), Gaps = 4/32 (12%)
Query 16 CTVYVLSLSWVVLFSLFSV----FNKCKDAVA 43
C VY + L+WV +LFS+ FN +DA++
Sbjct 125 CVVYTILLTWVAELALFSIFYHYFNGYEDAIS 156
> Hs20542893
Length=804
Score = 29.6 bits (65), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 82 CLVKQNLSVLDLVSVIMRRDNYIIALTNKEVLGRALPL 119
C+ + L+ LD+ I+R NY +AL NK +L PL
Sbjct 296 CVQPRPLTELDIHHRILRYTNYQVALANKGLLPARCPL 333
> CE17202
Length=899
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 23/76 (30%), Positives = 41/76 (53%), Gaps = 7/76 (9%)
Query 10 GPWQTL--CTVY--VLSLSWVVLFSLF--SVFNKCKDAVALRLYFRSRLYIHCDTALQLM 63
GP T+ C ++ VL L WV + L+ ++N C++ + + + LY+ C TAL+++
Sbjct 411 GPPPTIIECAIFLWVLGLIWVEIKQLWECGLYNYCRNLWNILDFITNSLYL-CTTALRVV 469
Query 64 GWPEVAALLLRAQQQH 79
+ +V LRA H
Sbjct 470 AYVQVEQEALRANSVH 485
> CE20100
Length=880
Score = 28.5 bits (62), Expect = 2.9, Method: Composition-based stats.
Identities = 13/57 (22%), Positives = 28/57 (49%), Gaps = 0/57 (0%)
Query 57 DTALQLMGWPEVAALLLRAQQQHPFCLVKQNLSVLDLVSVIMRRDNYIIALTNKEVL 113
D L + W + + AQ++ + + N++ + + I+R NY+ + NK +L
Sbjct 260 DDQLPNLTWHAIVKRICEAQKKLRLSIHQDNITSIYIYHRILRYKNYMTGMINKRIL 316
> CE13227
Length=779
Score = 28.1 bits (61), Expect = 3.9, Method: Composition-based stats.
Identities = 22/58 (37%), Positives = 27/58 (46%), Gaps = 9/58 (15%)
Query 14 TLCTVYVLSLSWVVLFSLFSVFNKCKDAV-----ALRLY----FRSRLYIHCDTALQL 62
T C V+ LSW VL SL ++F D + L LY F S+ YI C A L
Sbjct 581 TYCIVFTGVLSWTVLTSLNNLFRLLPDFLLRFIQPLLLYSSDLFWSKSYIFCGLAFAL 638
> At1g77000
Length=360
Score = 27.7 bits (60), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 21/34 (61%), Gaps = 0/34 (0%)
Query 87 NLSVLDLVSVIMRRDNYIIALTNKEVLGRALPLY 120
+L LDL S ++ D ++AL N+ + R+L LY
Sbjct 223 DLRTLDLCSCVLITDESVVALANRCIHLRSLGLY 256
> Hs20555642
Length=262
Score = 27.3 bits (59), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 19/77 (24%), Positives = 34/77 (44%), Gaps = 6/77 (7%)
Query 7 SPWGPWQTLCTVYVLSLSWVVLFSLFSVFNKCKDAVALRLYFRSRLYIHCDTALQLMGWP 66
PW W L + +SW+++FS+ VF+ + L ++Y H + +
Sbjct 131 EPWATWVPLLCFVLHVISWLLIFSILLVFDYAE------LMGLKQVYYHVLGLGEPLALK 184
Query 67 EVAALLLRAQQQHPFCL 83
AL L + +HP C+
Sbjct 185 SPRALRLFSHLRHPVCV 201
Lambda K H
0.330 0.141 0.459
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1209785478
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40