bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1740_orf1
Length=135
Score E
Sequences producing significant alignments: (Bits) Value
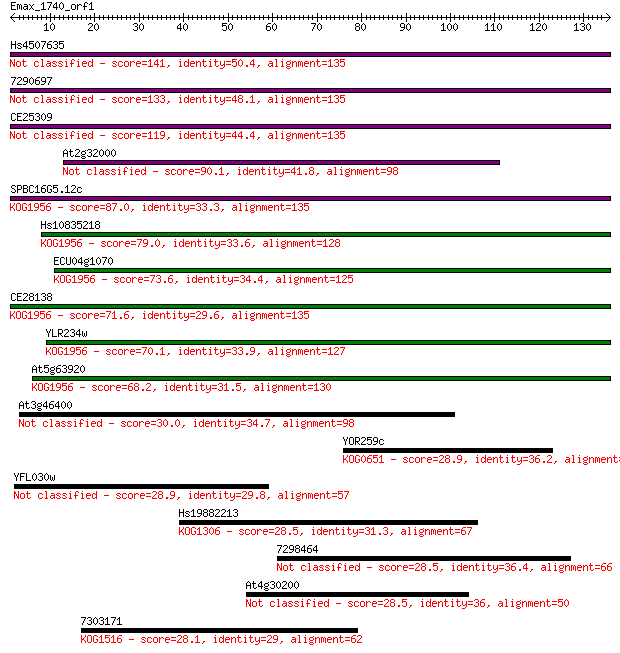
Hs4507635 141 4e-34
7290697 133 9e-32
CE25309 119 2e-27
At2g32000 90.1 1e-18
SPBC16G5.12c 87.0 9e-18
Hs10835218 79.0 2e-15
ECU04g1070 73.6 1e-13
CE28138 71.6 4e-13
YLR234w 70.1 1e-12
At5g63920 68.2 4e-12
At3g46400 30.0 1.1
YOR259c 28.9 2.6
YFL030w 28.9 2.7
Hs19882213 28.5 3.9
7298464 28.5 3.9
At4g30200 28.5 4.2
7303171 28.1 5.4
> Hs4507635
Length=862
Score = 141 bits (355), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 68/136 (50%), Positives = 90/136 (66%), Gaps = 1/136 (0%)
Query 1 FQGKYGDLDASLLSYGPCQTPTLQLCVHRSDAINSFTPETYYYIDAAVSVG-GQEIHMNW 59
FQGKYGDLD+SL+S+GPCQTPTL CV R D I SF PETY+ + A V+ + + ++W
Sbjct 200 FQGKYGDLDSSLISFGPCQTPTLGFCVERHDKIQSFKPETYWVLQAKVNTDKDRSLLLDW 259
Query 60 SRRQVFDLSVAQTFLAIVSEGIGGKCESIKETEEIICMPKALNTVELLKAASNRLGIGPH 119
R +VFD +AQ FL + + E+ E+ P ALNTVE+L+ AS+ LG+GP
Sbjct 260 DRVRVFDREIAQMFLNMTKLEKEAQVEATSRKEKAKQRPLALNTVEMLRVASSSLGMGPQ 319
Query 120 RAMQIAERLYLGGFIT 135
AMQ AERLY G+I+
Sbjct 320 HAMQTAERLYTQGYIS 335
> 7290697
Length=875
Score = 133 bits (334), Expect = 9e-32, Method: Compositional matrix adjust.
Identities = 65/135 (48%), Positives = 88/135 (65%), Gaps = 2/135 (1%)
Query 1 FQGKYGDLDASLLSYGPCQTPTLQLCVHRSDAINSFTPETYYYIDAAVSVGGQEIHMNWS 60
FQ +YGDLD+SL+SYGPCQTPTL CV R D I +F PE+++++ + G E+ + W+
Sbjct 199 FQDRYGDLDSSLISYGPCQTPTLGFCVKRHDDIQTFKPESFWHLQ--LLAGQPEVTLEWA 256
Query 61 RRQVFDLSVAQTFLAIVSEGIGGKCESIKETEEIICMPKALNTVELLKAASNRLGIGPHR 120
R +VF +A L V E ES+ E P+ALNTVEL++ S+ LGIGP +
Sbjct 257 RGRVFKKDIAIMLLNRVKEHKKATVESVASKEAYKSKPQALNTVELMRICSSGLGIGPFQ 316
Query 121 AMQIAERLYLGGFIT 135
AMQIAERLY G+I+
Sbjct 317 AMQIAERLYTQGYIS 331
> CE25309
Length=615
Score = 119 bits (298), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 60/137 (43%), Positives = 82/137 (59%), Gaps = 2/137 (1%)
Query 1 FQGKYGDLDASLLSYGPCQTPTLQLCVHRSDAINSFTPETYYYI--DAAVSVGGQEIHMN 58
FQGKYGDLD++++SYGPCQTPTL CV R D I F PE Y+ + + S GQ
Sbjct 201 FQGKYGDLDSNVISYGPCQTPTLGFCVTRHDQIVQFKPEQYWVLKTNFTTSDDGQIFSPE 260
Query 59 WSRRQVFDLSVAQTFLAIVSEGIGGKCESIKETEEIICMPKALNTVELLKAASNRLGIGP 118
W R ++FD VA+ F+ + + G + + E P ALNTVEL++ AS+ LG+ P
Sbjct 261 WQRGRIFDAEVARVFVDRIKKCHTGLVLDVSKKEARKERPCALNTVELMRVASSSLGMSP 320
Query 119 HRAMQIAERLYLGGFIT 135
M +AE LY G+I+
Sbjct 321 STTMHVAEALYTQGYIS 337
> At2g32000
Length=836
Score = 90.1 bits (222), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 41/98 (41%), Positives = 55/98 (56%), Gaps = 0/98 (0%)
Query 13 LSYGPCQTPTLQLCVHRSDAINSFTPETYYYIDAAVSVGGQEIHMNWSRRQVFDLSVAQT 72
YGPCQTPTL CV R IN+F PE ++ + + G E+ + W RR++FDL A
Sbjct 227 FRYGPCQTPTLGFCVQRYMHINTFKPEKFWALRPYIRKDGYELQLEWERRRLFDLEAATV 286
Query 73 FLAIVSEGIGGKCESIKETEEIICMPKALNTVELLKAA 110
F +V EG K + E +E+ P LNTV LLK +
Sbjct 287 FQKLVVEGRTAKVMDVSEKQEVKGRPAGLNTVNLLKCS 324
> SPBC16G5.12c
Length=622
Score = 87.0 bits (214), Expect = 9e-18, Method: Compositional matrix adjust.
Identities = 45/135 (33%), Positives = 64/135 (47%), Gaps = 0/135 (0%)
Query 1 FQGKYGDLDASLLSYGPCQTPTLQLCVHRSDAINSFTPETYYYIDAAVSVGGQEIHMNWS 60
Q + L ++SYGPCQ PTL V R + F PETY+++ G+ I NW
Sbjct 195 LQKSFDILQNKIISYGPCQFPTLGFVVDRWQRVEDFVPETYWHLRFVDKRQGKTIQFNWE 254
Query 61 RRQVFDLSVAQTFLAIVSEGIGGKCESIKETEEIICMPKALNTVELLKAASNRLGIGPHR 120
R +VFD L E K +I + + P L+TVEL K L I +
Sbjct 255 RAKVFDRLTTMIILENCLECKTAKVVNITQKPKTKYKPLPLSTVELTKLGPKHLRISAKK 314
Query 121 AMQIAERLYLGGFIT 135
+++AE LY GF++
Sbjct 315 TLELAENLYTNGFVS 329
> Hs10835218
Length=1001
Score = 79.0 bits (193), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 43/128 (33%), Positives = 63/128 (49%), Gaps = 0/128 (0%)
Query 8 LDASLLSYGPCQTPTLQLCVHRSDAINSFTPETYYYIDAAVSVGGQEIHMNWSRRQVFDL 67
L L+SYG CQ PTL V R AI +F PE ++ I + NW R ++F+
Sbjct 234 LAEQLISYGSCQFPTLGFVVERFKAIQAFVPEIFHRIKVTHDHKDGIVEFNWKRHRLFNH 293
Query 68 SVAQTFLAIVSEGIGGKCESIKETEEIICMPKALNTVELLKAASNRLGIGPHRAMQIAER 127
+ + E ++ + P+AL+TVEL K AS +L I M+IAE+
Sbjct 294 TACLVLYQLCVEDPMATVVEVRSKPKSKWRPQALDTVELEKLASRKLRINAKETMRIAEK 353
Query 128 LYLGGFIT 135
LY G+I+
Sbjct 354 LYTQGYIS 361
> ECU04g1070
Length=621
Score = 73.6 bits (179), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 43/125 (34%), Positives = 61/125 (48%), Gaps = 1/125 (0%)
Query 11 SLLSYGPCQTPTLQLCVHRSDAINSFTPETYYYIDAAVSVGGQEIHMNWSRRQVFDLSVA 70
+LS+GPCQ PTL V R+ SF PE ++ + G + W R VFD +
Sbjct 92 KILSFGPCQVPTLGFVVERARERESFVPEIFWTLKFKAMHKGFKDEFTWRRGPVFDRNCV 151
Query 71 QTFLAIVSEGIGGKCESIKETEEIICMPKALNTVELLKAASNRLGIGPHRAMQIAERLYL 130
F +S G + S + E+ P L TVEL K S+ + H+ M+IAE LY
Sbjct 152 VCFFNSLS-GEDARIVSREVKEKTKYRPLPLRTVELQKKCSSYFRLSGHKVMEIAEGLYN 210
Query 131 GGFIT 135
G+I+
Sbjct 211 RGYIS 215
> CE28138
Length=759
Score = 71.6 bits (174), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 40/136 (29%), Positives = 64/136 (47%), Gaps = 1/136 (0%)
Query 1 FQGKYGDLDAS-LLSYGPCQTPTLQLCVHRSDAINSFTPETYYYIDAAVSVGGQEIHMNW 59
F+ G D S ++SYG CQ PTL R I +F E ++ + + ++ W
Sbjct 198 FRDLLGQNDTSQVISYGSCQFPTLGFVTDRYKMIENFVSEPFWKLIVEHTRESHKVEFLW 257
Query 60 SRRQVFDLSVAQTFLAIVSEGIGGKCESIKETEEIICMPKALNTVELLKAASNRLGIGPH 119
R ++FD E E + + + P+AL+TVEL K ++L +
Sbjct 258 DRNRLFDRDTVDILHDECKETKEAHVEKVAKKPKSKWRPQALDTVELEKLGISKLRMSAK 317
Query 120 RAMQIAERLYLGGFIT 135
+ MQ+AE+LY GFI+
Sbjct 318 QTMQVAEKLYSKGFIS 333
> YLR234w
Length=656
Score = 70.1 bits (170), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 43/131 (32%), Positives = 60/131 (45%), Gaps = 4/131 (3%)
Query 9 DASLLSYGPCQTPTLQLCVHRSDAINSFTPETYYYIDAAV--SVGGQEIHMNWSRRQVFD 66
D+ ++SYG CQ PTL V R + I +F PE ++YI V G W R +FD
Sbjct 225 DSQVVSYGTCQFPTLGFVVDRFERIRNFVPEEFWYIQLVVENKDNGGTTTFQWDRGHLFD 284
Query 67 LSVAQTFLAIVSEGIGGKCE--SIKETEEIICMPKALNTVELLKAASNRLGIGPHRAMQI 124
TF E G + +K P L TVEL K + L + +++
Sbjct 285 RLSVLTFYETCIETAGNVAQVVDLKSKPTTKYRPLPLTTVELQKNCARYLRLNAKQSLDA 344
Query 125 AERLYLGGFIT 135
AE+LY GFI+
Sbjct 345 AEKLYQKGFIS 355
> At5g63920
Length=926
Score = 68.2 bits (165), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 41/131 (31%), Positives = 60/131 (45%), Gaps = 1/131 (0%)
Query 6 GDLDASLLSYGPCQTPTLQLCVHRSDAINSFTPETYYYIDAAVSVGGQEIHMNWSRRQVF 65
G+ + ++SYGPCQ PTL V R I + PE ++ I+ + NW R +F
Sbjct 211 GEERSRVISYGPCQFPTLGFIVERYWEIQAHEPEEFWTINCSHQSEEGLATFNWMRGHLF 270
Query 66 DLSVAQTFLAIVSEGIGGKCESIKETEEIICMPK-ALNTVELLKAASNRLGIGPHRAMQI 124
D + A + E ++ E P LNT+EL K AS + M++
Sbjct 271 DYASAVILYEMCVEEPTATVMNVPHPRERFKYPPYPLNTIELEKRASRYFRLSSEHTMKV 330
Query 125 AERLYLGGFIT 135
AE LY GFI+
Sbjct 331 AEELYQAGFIS 341
> At3g46400
Length=867
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 34/105 (32%), Positives = 48/105 (45%), Gaps = 18/105 (17%)
Query 3 GKYGDLDASLLSYGPCQTPTLQLCVHRSDAI-NSFTPETY----YYIDAAVSVGGQEIHM 57
GK G +DASL S P TL+ + D I N + Y Y I A ++ G + +
Sbjct 62 GKIGRIDASLESKYPRSQTTLR---YFPDGIRNCYNVNVYKGTNYLIRATINYGNYD-GL 117
Query 58 NWSRRQVFDLSVAQTFLAIVS--EGIGGKCESIKETEEIICMPKA 100
N S R FDL + F + + +GG EEII +PK+
Sbjct 118 NISPR--FDLYIGPNFWVTIDLEKHVGGDT-----WEEIIHIPKS 155
> YOR259c
Length=437
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 26/47 (55%), Gaps = 6/47 (12%)
Query 76 IVSEGIGGKCESIKETEEIICMPKALNTVELLKAASNRLGIGPHRAM 122
I +GIGG E I+E E+I +P L E+ + R+GI P + +
Sbjct 178 ITFDGIGGLTEQIRELREVIELP--LKNPEIFQ----RVGIKPPKGV 218
> YFL030w
Length=385
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 27/57 (47%), Gaps = 3/57 (5%)
Query 2 QGKYGDLDASLLSYGPCQTPTLQLCVHRSDAINSFTPETYYYIDAAVSVGGQEIHMN 58
Q YG A +++ T L S AI +PET++ +DA S+G +E +
Sbjct 135 QNSYG---AVTVTHVDTSTAVLSDLKAISQAIKQTSPETFFVVDAVCSIGCEEFEFD 188
> Hs19882213
Length=6307
Score = 28.5 bits (62), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 21/69 (30%), Positives = 34/69 (49%), Gaps = 3/69 (4%)
Query 39 ETYYYID-AAVSVGG-QEIHMNWSRRQVFDLSVAQTFLAIVSEGIGGKCESIKETEEIIC 96
E ++YI+ +V + G Q+ +NWS R D SVA + ++ + G S ET +
Sbjct 5088 EEFFYINLTSVEIRGLQKFDVNWSPRLNLDFSVA-VITILDNDDLAGMDISFPETTVAVA 5146
Query 97 MPKALNTVE 105
+ L VE
Sbjct 5147 VDTTLIPVE 5155
> 7298464
Length=2091
Score = 28.5 bits (62), Expect = 3.9, Method: Composition-based stats.
Identities = 24/69 (34%), Positives = 35/69 (50%), Gaps = 4/69 (5%)
Query 61 RRQVFDLSVAQTFLAIVSEGIGGKCESIKETEEIIC-MPKALNTVELLKA--ASNRLGIG 117
+R+ L + + LAI+ +G GKC E +E +C KA V+ L A A R I
Sbjct 1769 QRRDLHLELLRRKLAIIEDGARGKCMLQGERDEAVCRAKKAAKQVDKLSAQLADARSQIA 1828
Query 118 PHRAMQIAE 126
+A Q+AE
Sbjct 1829 EVKA-QLAE 1836
> At4g30200
Length=714
Score = 28.5 bits (62), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 18/67 (26%), Positives = 28/67 (41%), Gaps = 19/67 (28%)
Query 54 EIHMNWSRRQVFDL-----------------SVAQTFLAIVSEGIGGKCESIKETEEIIC 96
E+ WSR+++ + + +T L IVSE G+CE K + C
Sbjct 39 EVLQAWSRQEILQILCAEMGKERKYTGLTKVKIIETLLKIVSEKNSGECEGKKRDSD--C 96
Query 97 MPKALNT 103
+P NT
Sbjct 97 LPIQRNT 103
> 7303171
Length=800
Score = 28.1 bits (61), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 32/65 (49%), Gaps = 7/65 (10%)
Query 17 PCQTPTLQLCVHRSDAINSFTPET---YYYIDAAVSVGGQEIHMNWSRRQVFDLSVAQTF 73
P T + C++ +N +TPE+ Y + V V G+E +W R ++ L +A
Sbjct 109 PRSVSTDEDCLY----LNIWTPESGMRYGKLPIVVIVTGEEFAYDWPRNRINGLDLAGEG 164
Query 74 LAIVS 78
+ +VS
Sbjct 165 IVVVS 169
Lambda K H
0.321 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1388795348
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40