bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1822_orf1
Length=159
Score E
Sequences producing significant alignments: (Bits) Value
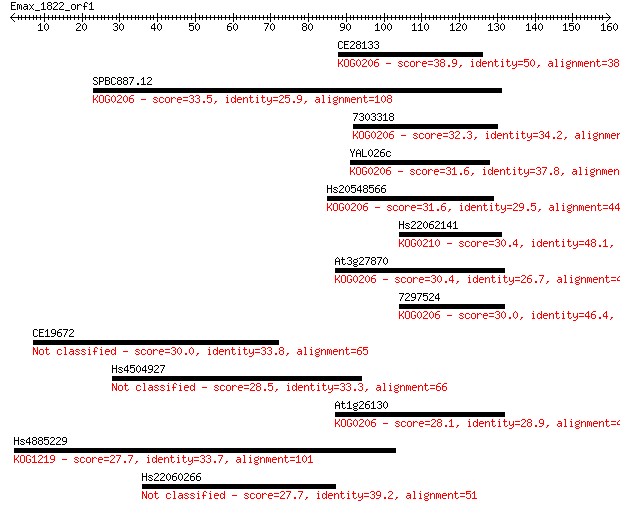
CE28133 38.9 0.004
SPBC887.12 33.5 0.19
7303318 32.3 0.42
YAL026c 31.6 0.67
Hs20548566 31.6 0.73
Hs22062141 30.4 1.3
At3g27870 30.4 1.4
7297524 30.0 1.9
CE19672 30.0 2.1
Hs4504927 28.5 5.1
At1g26130 28.1 7.7
Hs4885229 27.7 8.6
Hs22060266 27.7 9.3
> CE28133
Length=1139
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/38 (50%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 88 FYDFQIFEDLSVGDERSHRVNEFLKCMALCNTVVPHYK 125
F D + ED GDE S + E LK MA+C+TVVP K
Sbjct 418 FADASLIEDYRQGDEHSTSILEVLKMMAVCHTVVPENK 455
> SPBC887.12
Length=1258
Score = 33.5 bits (75), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 28/114 (24%), Positives = 49/114 (42%), Gaps = 8/114 (7%)
Query 23 EAVETPCSSVLSS-SHSLGASGGGNSFEGTTNGDSRKSVSFLYDSISPPTQFRETPADGQ 81
E +TP + SS LG G F T +R + F +I+ P D Q
Sbjct 492 EETDTPAACRTSSLVEELGQVG--YIFSDKTGTLTRNQMEFRQCTIAGVAYADVIPEDRQ 549
Query 82 -----IRRNCNFYDFQIFEDLSVGDERSHRVNEFLKCMALCNTVVPHYKANNNT 130
+ + YDF ++ E + +++FL +++C+TV+P Y + N+
Sbjct 550 FTSEDLDSDMYIYDFDTLKENLKHSENASLIHQFLLVLSICHTVIPEYDESTNS 603
> 7303318
Length=1297
Score = 32.3 bits (72), Expect = 0.42, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 92 QIFEDLSVGDERSHRVNEFLKCMALCNTVVPHYKANNN 129
Q+ +++ E S + EFL+ +++C+TV+P K N N
Sbjct 523 QLVQNILGRHETSAVIEEFLELLSVCHTVIPERKENGN 560
> YAL026c
Length=1355
Score = 31.6 bits (70), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 26/42 (61%), Gaps = 5/42 (11%)
Query 91 FQIFEDLSV-----GDERSHRVNEFLKCMALCNTVVPHYKAN 127
++ F+DL DE S +N+FL +A C+TV+P ++++
Sbjct 602 YRKFDDLKKKLNDPSDEDSPIINDFLTLLATCHTVIPEFQSD 643
> Hs20548566
Length=721
Score = 31.6 bits (70), Expect = 0.73, Method: Composition-based stats.
Identities = 13/44 (29%), Positives = 26/44 (59%), Gaps = 0/44 (0%)
Query 85 NCNFYDFQIFEDLSVGDERSHRVNEFLKCMALCNTVVPHYKANN 128
+C+F D ++ +++ + + EFL +A+C+TVVP +N
Sbjct 8 SCDFDDPRLLKNIEDRHPTAPCIQEFLTLLAVCHTVVPEKDGDN 51
> Hs22062141
Length=875
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 13/27 (48%), Positives = 19/27 (70%), Gaps = 0/27 (0%)
Query 104 SHRVNEFLKCMALCNTVVPHYKANNNT 130
S RV+E +K +ALC+ V P Y++N T
Sbjct 281 SSRVHEAVKAIALCHNVTPVYESNGVT 307
> At3g27870
Length=1174
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 12/45 (26%), Positives = 26/45 (57%), Gaps = 2/45 (4%)
Query 87 NFYDFQIFEDLSVGDERSHRVNEFLKCMALCNTVVPHYKANNNTG 131
NF+D +I + + + + +F + +A+C+T +P N++TG
Sbjct 487 NFWDERIVDGQWINQPNAELIQKFFRVLAICHTAIP--DVNSDTG 529
> 7297524
Length=1148
Score = 30.0 bits (66), Expect = 1.9, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 19/32 (59%), Gaps = 4/32 (12%)
Query 104 SHRVNEFLKCMALCNTVV----PHYKANNNTG 131
+ R+ EFL +A+CNTV+ PH N +G
Sbjct 361 AQRIQEFLVVLAICNTVIVGAAPHRDMMNASG 392
> CE19672
Length=279
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 30/65 (46%), Gaps = 8/65 (12%)
Query 7 AFAVEDDVMESSPEESEAVETPCSSVLSSSHSLGASGGGNSFEGTTNGDSRKSVSFLYDS 66
+F EDD +E E E T C V SS ++ + +S SR V FL+D
Sbjct 52 SFLYEDDDVEELVERDELSRTYCP-VCSSRDTVPLNFISHSL-------SRLQVKFLFDL 103
Query 67 ISPPT 71
+ PPT
Sbjct 104 LMPPT 108
> Hs4504927
Length=467
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 22/71 (30%), Positives = 35/71 (49%), Gaps = 6/71 (8%)
Query 28 PCSSVLSSSHSLGASGGGNSF-EGTTNGDSRKSVSFLYDSIS----PPTQFRETPADGQI 82
P S + S H+ G G G F EG+ NG ++++ FL D ++ Q A+ +
Sbjct 64 PGSYLSSECHTSGFVGSGGWFCEGSFNGSEKETMQFLNDRLANYLEKVRQLERENAELES 123
Query 83 RRNCNFYDFQI 93
R +Y+FQI
Sbjct 124 RIQ-EWYEFQI 133
> At1g26130
Length=1184
Score = 28.1 bits (61), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 13/45 (28%), Positives = 27/45 (60%), Gaps = 2/45 (4%)
Query 87 NFYDFQIFEDLSVGDERSHRVNEFLKCMALCNTVVPHYKANNNTG 131
NF D +I + V + + + +F + +A+C+TV+P + + +TG
Sbjct 494 NFRDERIMDGNWVTETHADVIQKFFQLLAVCHTVIP--EVDEDTG 536
> Hs4885229
Length=4590
Score = 27.7 bits (60), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 34/107 (31%), Positives = 47/107 (43%), Gaps = 18/107 (16%)
Query 2 SKKNIAFAVEDDVMESSPEESEAVETPCSSVLSSSHSLGASGGGNSF-----EGTTNGD- 55
S +I +A+E D ES E E S V+++ SL G N F NG
Sbjct 2629 SNADITYAIEAD-SESVKENLEI--NKLSGVITTKESL--IGLENEFFTFFVRAVDNGSP 2683
Query 56 SRKSVSFLYDSISPPTQFRETPADGQIRRNCNFYDFQIFEDLSVGDE 102
S++SV +Y I PP + P + FY F + ED+ VG E
Sbjct 2684 SKESVVLVYVKILPPEM--QLPKFSE-----PFYTFTVSEDVPVGTE 2723
> Hs22060266
Length=531
Score = 27.7 bits (60), Expect = 9.3, Method: Composition-based stats.
Identities = 20/51 (39%), Positives = 26/51 (50%), Gaps = 3/51 (5%)
Query 36 SHSLGASGGGNSFEGTTNGDSRKSVSFLYDSISPPTQFRETPADGQIRRNC 86
SH LGA G G+S + T D K S L +++SP +T DG R C
Sbjct 146 SHRLGAGGLGSSNQAVT--DLMKMDSPLQEALSPDAS-TQTALDGASWRKC 193
Lambda K H
0.312 0.129 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2136300674
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40