bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
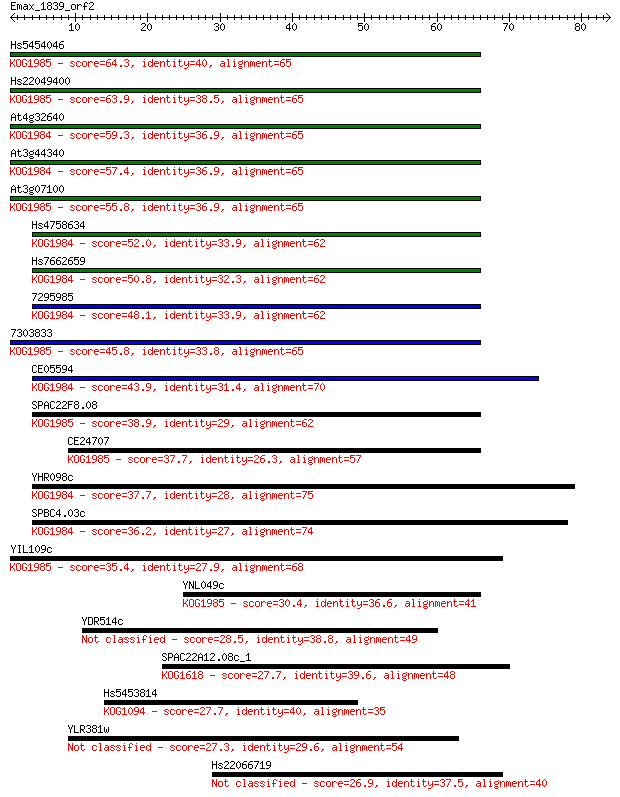
Query= Emax_1839_orf2
Length=83
Score E
Sequences producing significant alignments: (Bits) Value
Hs5454046 64.3 5e-11
Hs22049400 63.9 6e-11
At4g32640 59.3 2e-09
At3g44340 57.4 7e-09
At3g07100 55.8 2e-08
Hs4758634 52.0 3e-07
Hs7662659 50.8 7e-07
7295985 48.1 4e-06
7303833 45.8 2e-05
CE05594 43.9 7e-05
SPAC22F8.08 38.9 0.002
CE24707 37.7 0.005
YHR098c 37.7 0.005
SPBC4.03c 36.2 0.017
YIL109c 35.4 0.024
YNL049c 30.4 0.95
YDR514c 28.5 3.0
SPAC22A12.08c_1 27.7 6.1
Hs5453814 27.7 6.1
YLR381w 27.3 7.8
Hs22066719 26.9 9.4
> Hs5454046
Length=1268
Score = 64.3 bits (155), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 45/65 (69%), Gaps = 0/65 (0%)
Query 1 TLAAVLTYDSSVHFYRLEASQTRPRLFVVPQVQDIFLPLAENLFVPLHECKEAILNTLNA 60
T +T+DS++HFY L+ ++P++ +V + D+FLP ++L V L+E KE I + LNA
Sbjct 716 TRIGFMTFDSTIHFYNLQEGLSQPQMLIVSDIDDVFLPTPDSLLVNLYESKELIKDLLNA 775
Query 61 IPDIF 65
+P++F
Sbjct 776 LPNMF 780
> Hs22049400
Length=1093
Score = 63.9 bits (154), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 45/65 (69%), Gaps = 0/65 (0%)
Query 1 TLAAVLTYDSSVHFYRLEASQTRPRLFVVPQVQDIFLPLAENLFVPLHECKEAILNTLNA 60
T +T+DS++HFY L+ S ++P++ +V ++D+F+P+ ENL V L+E KE + + L
Sbjct 542 TKIGFITFDSTIHFYGLQESLSQPQMLIVSDIEDVFIPMPENLLVNLNESKELVQDLLKT 601
Query 61 IPDIF 65
+P +F
Sbjct 602 LPQMF 606
> At4g32640
Length=1069
Score = 59.3 bits (142), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 43/65 (66%), Gaps = 0/65 (0%)
Query 1 TLAAVLTYDSSVHFYRLEASQTRPRLFVVPQVQDIFLPLAENLFVPLHECKEAILNTLNA 60
T + T+DS++HFY L+ + +P + +VP VQD++ PL ++ V L EC++ + L++
Sbjct 543 TFVGIATFDSTIHFYNLKRALQQPLMLIVPDVQDVYTPLETDVVVQLSECRQHLELLLDS 602
Query 61 IPDIF 65
IP +F
Sbjct 603 IPTMF 607
> At3g44340
Length=1122
Score = 57.4 bits (137), Expect = 7e-09, Method: Composition-based stats.
Identities = 24/65 (36%), Positives = 42/65 (64%), Gaps = 0/65 (0%)
Query 1 TLAAVLTYDSSVHFYRLEASQTRPRLFVVPQVQDIFLPLAENLFVPLHECKEAILNTLNA 60
T + T+DS++HFY L+ + +P + +VP VQD++ PL ++ V L EC++ + L +
Sbjct 557 TFVGIATFDSTIHFYNLKRALQQPLMLIVPDVQDVYTPLETDVIVQLSECRQHLEILLES 616
Query 61 IPDIF 65
IP +F
Sbjct 617 IPTMF 621
> At3g07100
Length=1054
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 24/65 (36%), Positives = 44/65 (67%), Gaps = 0/65 (0%)
Query 1 TLAAVLTYDSSVHFYRLEASQTRPRLFVVPQVQDIFLPLAENLFVPLHECKEAILNTLNA 60
T +TYDS++HFY +++S ++P++ VV + DIF+PL ++L V L E + + L++
Sbjct 482 TQIGFITYDSTLHFYNMKSSLSQPQMMVVSDLDDIFVPLPDDLLVNLSESRTVVDAFLDS 541
Query 61 IPDIF 65
+P +F
Sbjct 542 LPLMF 546
> Hs4758634
Length=1125
Score = 52.0 bits (123), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 40/62 (64%), Gaps = 0/62 (0%)
Query 4 AVLTYDSSVHFYRLEASQTRPRLFVVPQVQDIFLPLAENLFVPLHECKEAILNTLNAIPD 63
+TY+ +HFY +++S +P++ VV V D+F+PL + V ++E + I + L+ IP+
Sbjct 548 GFVTYNKVLHFYNVKSSLAQPQMMVVSDVADMFVPLLDGFLVNVNESRAVITSLLDQIPE 607
Query 64 IF 65
+F
Sbjct 608 MF 609
> Hs7662659
Length=1032
Score = 50.8 bits (120), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 20/62 (32%), Positives = 38/62 (61%), Gaps = 0/62 (0%)
Query 4 AVLTYDSSVHFYRLEASQTRPRLFVVPQVQDIFLPLAENLFVPLHECKEAILNTLNAIPD 63
+TY+ +HF+ ++++ +P++ VV V ++F+PL + V E + I N L+ IPD
Sbjct 486 GFITYNKVLHFFNVKSNLAQPQMMVVTDVGEVFVPLLDGFLVNYQESQSVIHNLLDQIPD 545
Query 64 IF 65
+F
Sbjct 546 MF 547
> 7295985
Length=1192
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 36/62 (58%), Gaps = 0/62 (0%)
Query 4 AVLTYDSSVHFYRLEASQTRPRLFVVPQVQDIFLPLAENLFVPLHECKEAILNTLNAIPD 63
+TY+S+VHFY +++S +P++ VV VQ++F+PL + E I + IP
Sbjct 648 GFITYNSTVHFYNIKSSLAQPQMMVVGDVQEMFMPLLDGFLCHPEESAAVIDALMEEIPR 707
Query 64 IF 65
+F
Sbjct 708 MF 709
> 7303833
Length=1184
Score = 45.8 bits (107), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 34/65 (52%), Gaps = 0/65 (0%)
Query 1 TLAAVLTYDSSVHFYRLEASQTRPRLFVVPQVQDIFLPLAENLFVPLHECKEAILNTLNA 60
T + +DS VHFY + +P + ++D FLP ++L V L ECKE + + L
Sbjct 623 TQVGFICFDSFVHFYSMAEGLNQPHEMTLLDIEDPFLPRPDSLLVNLKECKELVRDLLKQ 682
Query 61 IPDIF 65
+P F
Sbjct 683 LPKRF 687
> CE05594
Length=1126
Score = 43.9 bits (102), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 40/70 (57%), Gaps = 3/70 (4%)
Query 4 AVLTYDSSVHFYRLEASQTRPRLFVVPQVQDIFLPLAENLFVPLHECKEAILNTLNAIPD 63
+ T+D +VHF+ + S P++ V+ VQ+ F+P+ + L +P +E + L IP
Sbjct 580 GLATFDQAVHFFDI--SSASPKMLVMSDVQEPFVPMVDGLLLPYNEALPGLRAALAEIPK 637
Query 64 IFFSKAKRTR 73
+ FS++K T
Sbjct 638 L-FSQSKTTE 646
> SPAC22F8.08
Length=926
Score = 38.9 bits (89), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/62 (29%), Positives = 32/62 (51%), Gaps = 0/62 (0%)
Query 4 AVLTYDSSVHFYRLEASQTRPRLFVVPQVQDIFLPLAENLFVPLHECKEAILNTLNAIPD 63
A + DS++HF+ + VV +++ FLP ++L + L EC++ I N L
Sbjct 372 AFIGVDSALHFFSVSPGAEEATQLVVSDLEEPFLPRNQDLLLNLRECRQGIENLLERFQS 431
Query 64 IF 65
+F
Sbjct 432 MF 433
> CE24707
Length=1024
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 15/57 (26%), Positives = 30/57 (52%), Gaps = 0/57 (0%)
Query 9 DSSVHFYRLEASQTRPRLFVVPQVQDIFLPLAENLFVPLHECKEAILNTLNAIPDIF 65
D +HF+ +++ P +V + D FLP +L VP+ + K+ I + +P+ +
Sbjct 452 DQCLHFFSFSSNKRYPNEMIVDDIDDAFLPSVTSLLVPMKKFKDTIRTFIKQLPEFY 508
> YHR098c
Length=929
Score = 37.7 bits (86), Expect = 0.005, Method: Composition-based stats.
Identities = 21/75 (28%), Positives = 37/75 (49%), Gaps = 1/75 (1%)
Query 4 AVLTYDSSVHFYRLEASQTRPRLFVVPQVQDIFLPLAENLFVPLHECKEAILNTLNAIPD 63
A++ YD+ + F+ L + ++V ++ D+FLP LFV + I +TL I
Sbjct 339 AIIVYDNKLRFFNLRPDLDNAQEYIVSELDDVFLPFYNGLFVKPGNSMKIINDTLIKISG 398
Query 64 IFFSKAKRTRLPPPC 78
+ S K + +P C
Sbjct 399 -YISTDKYSHVPQVC 412
> SPBC4.03c
Length=891
Score = 36.2 bits (82), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 20/74 (27%), Positives = 38/74 (51%), Gaps = 4/74 (5%)
Query 4 AVLTYDSSVHFYRLEASQTRPRLFVVPQVQDIFLPLAENLFVPLHECKEAILNTLNAIPD 63
A++ +D SV+F+ L + +P + +++ F+P + LFV K I L++ P
Sbjct 334 AIVCFDRSVNFFNLSPNLEQPHMLAASDLENPFVPFSSGLFVDPIASKVVIERLLDSFPA 393
Query 64 IFFSKAKRTRLPPP 77
IF + ++P P
Sbjct 394 IF----QDVKIPEP 403
> YIL109c
Length=926
Score = 35.4 bits (80), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 19/75 (25%), Positives = 38/75 (50%), Gaps = 7/75 (9%)
Query 1 TLAAVLTYDSSVHFYRL-------EASQTRPRLFVVPQVQDIFLPLAENLFVPLHECKEA 53
T ++L D+++H++++ E S + + + +++ FLP ++ V L C++
Sbjct 343 TRISILCVDNAIHYFKIPLDSENNEESADQINMMDIADLEEPFLPRPNSMVVSLKACRQN 402
Query 54 ILNTLNAIPDIFFSK 68
I L IP IF S
Sbjct 403 IETLLTKIPQIFQSN 417
> YNL049c
Length=876
Score = 30.4 bits (67), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 25 RLFVVPQVQDIFLPL-AENLFVPLHECKEAILNTLNAIPDIF 65
++F + + + FLP+ ++ L VPL CK + L IP+IF
Sbjct 329 QMFDIGDLDEPFLPMPSDELVVPLKYCKNNLETLLKKIPEIF 370
> YDR514c
Length=483
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 19/49 (38%), Positives = 26/49 (53%), Gaps = 2/49 (4%)
Query 11 SVHFYRLEASQTRPRLFVVPQVQDIFLPLAENLFVPLHECKEAILNTLN 59
S H EA R + FV +D FL L E+L +PL +C E I + +N
Sbjct 239 SYHLIVAEALPLRNKKFVC-DFKDCFL-LGESLVLPLEQCVEFIQSLIN 285
> SPAC22A12.08c_1
Length=379
Score = 27.7 bits (60), Expect = 6.1, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 25/52 (48%), Gaps = 4/52 (7%)
Query 22 TRPRLFVVPQVQDIFLPLAENLFVPLHECKEAILNTL----NAIPDIFFSKA 69
TRP V V + L L L + L + K +L T+ N PDI+FS A
Sbjct 177 TRPIEAVFTYVDPVRLGLDLQLVMELGQSKNGVLGTVSKTANEGPDIYFSNA 228
> Hs5453814
Length=855
Score = 27.7 bits (60), Expect = 6.1, Method: Composition-based stats.
Identities = 14/35 (40%), Positives = 18/35 (51%), Gaps = 0/35 (0%)
Query 14 FYRLEASQTRPRLFVVPQVQDIFLPLAENLFVPLH 48
++R EAS+ P P V D P A + VPLH
Sbjct 305 YFRSEASEWEPNAISFPLVLDDVNPSARFVTVPLH 339
> YLR381w
Length=733
Score = 27.3 bits (59), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 16/54 (29%), Positives = 26/54 (48%), Gaps = 3/54 (5%)
Query 9 DSSVHFYRLEASQTRPRLFVVPQVQDIFLPLAENLFVPLHECKEAILNTLNAIP 62
D + + YR + ++ V P D F + ENL++P + K A T+ IP
Sbjct 587 DLTNYLYRNKVLSSKSLFGVSP---DFFKQILENLYIPTADFKNAKFFTITGIP 637
> Hs22066719
Length=361
Score = 26.9 bits (58), Expect = 9.4, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 29 VPQVQDIFLPLAENLFVPLHECKEAILNTLNAIPDIFFSK 68
VP+ D LP A P ++ ++A LN A+P FF K
Sbjct 206 VPRAHDPRLPEATPKGHPWNQAQQAPLNRSPALPSTFFGK 245
Lambda K H
0.325 0.138 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1200681374
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40