bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1861_orf1
Length=126
Score E
Sequences producing significant alignments: (Bits) Value
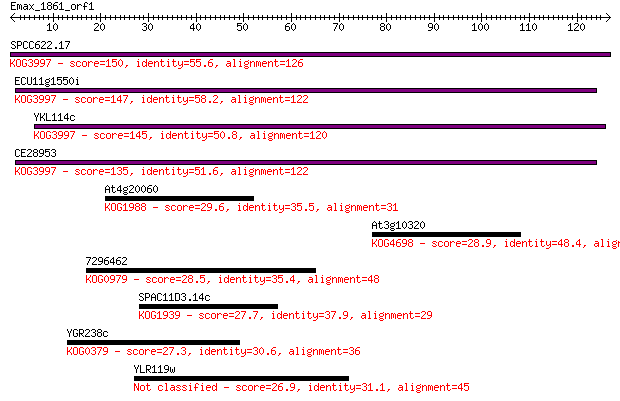
SPCC622.17 150 4e-37
ECU11g1550i 147 4e-36
YKL114c 145 1e-35
CE28953 135 2e-32
At4g20060 29.6 1.4
At3g10320 28.9 2.6
7296462 28.5 3.0
SPAC11D3.14c 27.7 6.0
YGR238c 27.3 7.0
YLR119w 26.9 8.0
> SPCC622.17
Length=366
Score = 150 bits (380), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 70/126 (55%), Positives = 92/126 (73%), Gaps = 0/126 (0%)
Query 1 SKEEGIRNIAAAVNRAHKDTKFVVVVLENMAGQKNVIGSKFEDLRDIIDLVEDKDRVGVC 60
+KEEGI N+A +NRAH++TK V++V ENMAGQ N +G F+D + +++ DR VC
Sbjct 133 TKEEGINNLAECINRAHEETKSVIIVTENMAGQGNCLGGTFDDFAALKSKIKNLDRWRVC 192
Query 61 LDTCHLFAAGYDIRTAEKFEKVMEDFDSIVGYKFLKGMHLNDSKSELSSGLDRHELLGKG 120
LDTCH FAAGYDIRT E ++KV+++FD VG K++ G HLNDSK+ L S D HE +G G
Sbjct 193 LDTCHTFAAGYDIRTEESYKKVIDEFDEKVGAKYVSGWHLNDSKAPLGSNRDLHENIGLG 252
Query 121 HLGLEP 126
LGLEP
Sbjct 253 FLGLEP 258
> ECU11g1550i
Length=275
Score = 147 bits (371), Expect = 4e-36, Method: Compositional matrix adjust.
Identities = 71/122 (58%), Positives = 91/122 (74%), Gaps = 0/122 (0%)
Query 2 KEEGIRNIAAAVNRAHKDTKFVVVVLENMAGQKNVIGSKFEDLRDIIDLVEDKDRVGVCL 61
+ E +R+I VN A + V V++ENMAGQ +V+GS FE+LRDII +ED++R+GVCL
Sbjct 113 RAECLRSIGHHVNTALSRVQGVTVLIENMAGQGSVVGSSFEELRDIIGGIEDRERIGVCL 172
Query 62 DTCHLFAAGYDIRTAEKFEKVMEDFDSIVGYKFLKGMHLNDSKSELSSGLDRHELLGKGH 121
DTCHLF AG+DI T EKF +VME FD +VG +FLK MH+NDSK L S DRHE +GKG
Sbjct 173 DTCHLFGAGFDITTEEKFLEVMERFDRVVGLEFLKAMHINDSKEALGSKKDRHESIGKGM 232
Query 122 LG 123
+G
Sbjct 233 IG 234
> YKL114c
Length=367
Score = 145 bits (367), Expect = 1e-35, Method: Compositional matrix adjust.
Identities = 61/120 (50%), Positives = 92/120 (76%), Gaps = 0/120 (0%)
Query 6 IRNIAAAVNRAHKDTKFVVVVLENMAGQKNVIGSKFEDLRDIIDLVEDKDRVGVCLDTCH 65
++ +A+ +N+A K+TKFV +VLENMAG N++GS DL+++I ++EDK R+GVC+DTCH
Sbjct 136 LKQLASYLNKAIKETKFVKIVLENMAGTGNLVGSSLVDLKEVIGMIEDKSRIGVCIDTCH 195
Query 66 LFAAGYDIRTAEKFEKVMEDFDSIVGYKFLKGMHLNDSKSELSSGLDRHELLGKGHLGLE 125
FAAGYDI T E F ++F+ ++G+K+L +HLNDSK+ L + D HE LG+G+LG++
Sbjct 196 TFAAGYDISTTETFNNFWKEFNDVIGFKYLSAVHLNDSKAPLGANRDLHERLGQGYLGID 255
> CE28953
Length=396
Score = 135 bits (339), Expect = 2e-32, Method: Composition-based stats.
Identities = 63/122 (51%), Positives = 91/122 (74%), Gaps = 0/122 (0%)
Query 2 KEEGIRNIAAAVNRAHKDTKFVVVVLENMAGQKNVIGSKFEDLRDIIDLVEDKDRVGVCL 61
KEE + IA ++ + T+ +++VLE MAGQ N IG FE+L+ IID V+ K RVGVC+
Sbjct 235 KEECMTTIAETIDFVVEKTENIILVLETMAGQGNSIGGTFEELKFIIDKVKVKSRVGVCI 294
Query 62 DTCHLFAAGYDIRTAEKFEKVMEDFDSIVGYKFLKGMHLNDSKSELSSGLDRHELLGKGH 121
DTCH+FA GYDIRT + +E+VM++F +VG+ +LK +H+NDSK ++ S LDRHE +G+G
Sbjct 295 DTCHIFAGGYDIRTQKAYEEVMKNFGEVVGWNYLKAIHINDSKGDVGSKLDRHEHIGQGK 354
Query 122 LG 123
+G
Sbjct 355 IG 356
> At4g20060
Length=1134
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 11/31 (35%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 21 KFVVVVLENMAGQKNVIGSKFEDLRDIIDLV 51
KF++V LEN+ G N++ +E ++ I + V
Sbjct 510 KFLIVFLENLEGDDNLLSEIYEKVKHITEFV 540
> At3g10320
Length=494
Score = 28.9 bits (63), Expect = 2.6, Method: Composition-based stats.
Identities = 15/36 (41%), Positives = 25/36 (69%), Gaps = 5/36 (13%)
Query 77 EKFEK---VMEDFDSIV--GYKFLKGMHLNDSKSEL 107
EK++K +++D +SI G++F KG++LND K L
Sbjct 441 EKYDKDDPILKDPNSITKKGWQFTKGIYLNDQKVRL 476
> 7296462
Length=1345
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 17/51 (33%), Positives = 28/51 (54%), Gaps = 7/51 (13%)
Query 17 HKDTKFVVVVLENMAGQKNVIGSKFEDLRDIIDLVED---KDRVGVCLDTC 64
H+D KF LEN+ Q+++ ED D+ DL+ + K ++GV + C
Sbjct 427 HEDAKF----LENVVAQRDLFAFACEDKGDMSDLINELCVKQKLGVNVIYC 473
> SPAC11D3.14c
Length=1260
Score = 27.7 bits (60), Expect = 6.0, Method: Composition-based stats.
Identities = 11/29 (37%), Positives = 21/29 (72%), Gaps = 0/29 (0%)
Query 28 ENMAGQKNVIGSKFEDLRDIIDLVEDKDR 56
EN + K+++ KF +LRDII++ +K++
Sbjct 413 ENESMNKDIVIEKFSELRDIINIDIEKEK 441
> YGR238c
Length=882
Score = 27.3 bits (59), Expect = 7.0, Method: Composition-based stats.
Identities = 11/36 (30%), Positives = 21/36 (58%), Gaps = 0/36 (0%)
Query 13 VNRAHKDTKFVVVVLENMAGQKNVIGSKFEDLRDII 48
+ A KD+ F L+N+ QK + S+ DL++++
Sbjct 591 IKEASKDSNFQTARLKNLEIQKTFLESRINDLKNLL 626
> YLR119w
Length=213
Score = 26.9 bits (58), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 14/46 (30%), Positives = 24/46 (52%), Gaps = 1/46 (2%)
Query 27 LENMAGQKNVIGSKFEDLRDII-DLVEDKDRVGVCLDTCHLFAAGY 71
L + AG+ + +F+ L + DL E KD+V L+ C + + Y
Sbjct 85 LTDFAGKIHAFRDQFKQLEENFEDLHEQKDKVQALLENCRILESKY 130
Lambda K H
0.319 0.138 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1180352192
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40