bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1872_orf1
Length=134
Score E
Sequences producing significant alignments: (Bits) Value
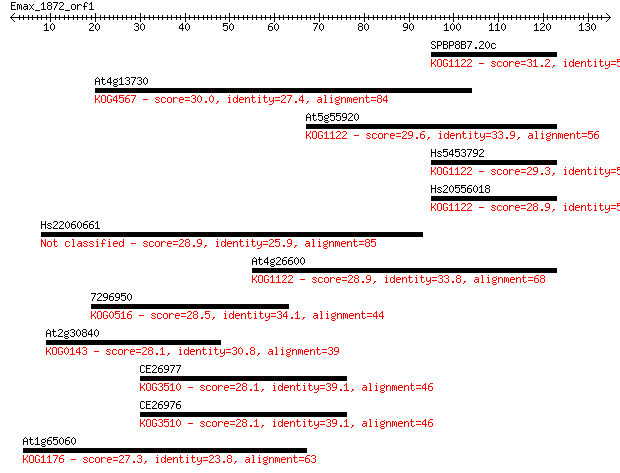
SPBP8B7.20c 31.2 0.56
At4g13730 30.0 1.4
At5g55920 29.6 1.6
Hs5453792 29.3 2.3
Hs20556018 28.9 2.7
Hs22060661 28.9 3.2
At4g26600 28.9 3.2
7296950 28.5 3.9
At2g30840 28.1 4.9
CE26977 28.1 5.0
CE26976 28.1 5.0
At1g65060 27.3 7.6
> SPBP8B7.20c
Length=608
Score = 31.2 bits (69), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 14/28 (50%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 95 PVYLRTNTLTITRDRLYHYLLNKGVAVE 122
PV +RTNTL R L L+N+GV +E
Sbjct 264 PVTIRTNTLKTQRRELAQALINRGVNLE 291
> At4g13730
Length=449
Score = 30.0 bits (66), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 23/93 (24%), Positives = 45/93 (48%), Gaps = 18/93 (19%)
Query 20 VKWKGLLDHITPPPANWPSRIRTYFVSDRWKLQAQNERLPASVRTSFPAELFRRIEASHG 79
+ WK LLD+++P + W S + K ++Q ++ + + P+E+ R+++ S G
Sbjct 134 IVWKLLLDYLSPDRSLWSSELA--------KKRSQYKQFKEELLMN-PSEVTRKMDKSKG 184
Query 80 TDK---------AIHICNILNEEAPVYLRTNTL 103
D A+ I +E+ P+ L T +L
Sbjct 185 GDSNDPKIESPGALSRSEITHEDHPLSLGTTSL 217
> At5g55920
Length=682
Score = 29.6 bits (65), Expect = 1.6, Method: Composition-based stats.
Identities = 19/56 (33%), Positives = 25/56 (44%), Gaps = 13/56 (23%)
Query 67 PAELFRRIEASHGTDKAIHICNILNEEAPVYLRTNTLTITRDRLYHYLLNKGVAVE 122
P EL IEA ++ P +RTNTL R L LLN+GV ++
Sbjct 269 PGELMELIEA-------------FEKQRPTSIRTNTLKTRRRDLADVLLNRGVNLD 311
> Hs5453792
Length=855
Score = 29.3 bits (64), Expect = 2.3, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 95 PVYLRTNTLTITRDRLYHYLLNKGVAVE 122
PV LRTNTL R L L+N+GV ++
Sbjct 309 PVTLRTNTLKTRRRDLAQALINRGVNLD 336
> Hs20556018
Length=812
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 14/28 (50%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 95 PVYLRTNTLTITRDRLYHYLLNKGVAVE 122
PV LRTNTL R L L+N+GV ++
Sbjct 309 PVTLRTNTLKTRRRDLAQALINRGVNLD 336
> Hs22060661
Length=388
Score = 28.9 bits (63), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 22/85 (25%), Positives = 37/85 (43%), Gaps = 11/85 (12%)
Query 8 DKKFITEHIYQIVKWKGLLDHITPPPANWPSRIRTYFVSDRWKLQAQNERLPASVRTSFP 67
+ +F T + V G+ +H++PPP PS I + R + E + +R +F
Sbjct 99 NNEFATRTPNKAVHVCGIYEHVSPPP---PSDIAMLMLPVRSFTVSAGEFPGSLLRKAFS 155
Query 68 AELFRRIEASHGTDKAIHICNILNE 92
EL R+ +HIC+ E
Sbjct 156 RELSSRLH--------VHICSFQGE 172
> At4g26600
Length=626
Score = 28.9 bits (63), Expect = 3.2, Method: Composition-based stats.
Identities = 23/69 (33%), Positives = 31/69 (44%), Gaps = 14/69 (20%)
Query 55 NERLPASVRTSFPA-ELFRRIEASHGTDKAIHICNILNEEAPVYLRTNTLTITRDRLYHY 113
NE L ++ FP EL IEA ++ P +RTNTL R L
Sbjct 239 NEFLIGTLIEMFPVVELMELIEA-------------FEKKRPTSIRTNTLKTRRRDLADI 285
Query 114 LLNKGVAVE 122
LLN+GV ++
Sbjct 286 LLNRGVNLD 294
> 7296950
Length=7182
Score = 28.5 bits (62), Expect = 3.9, Method: Composition-based stats.
Identities = 15/44 (34%), Positives = 21/44 (47%), Gaps = 0/44 (0%)
Query 19 IVKWKGLLDHITPPPANWPSRIRTYFVSDRWKLQAQNERLPASV 62
+ K K L DHI +N P+R + D L A E+ AS+
Sbjct 3673 VAKLKSLEDHIEQQASNIPARSKEVMARDLANLHADFEKFGASL 3716
> At2g30840
Length=362
Score = 28.1 bits (61), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 12/39 (30%), Positives = 18/39 (46%), Gaps = 0/39 (0%)
Query 9 KKFITEHIYQIVKWKGLLDHITPPPANWPSRIRTYFVSD 47
KKF T + + VK+ D + P ANW + + D
Sbjct 122 KKFYTRDVTKTVKYNSNFDLYSSPSANWRDTLSCFMAPD 160
> CE26977
Length=4450
Score = 28.1 bits (61), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 22/47 (46%), Gaps = 1/47 (2%)
Query 30 TPPPANWPSRIRTYFVSDRWKLQAQNERLPASVRT-SFPAELFRRIE 75
T P W +IRT V D K QA S+ T S P E+F +E
Sbjct 2997 TQPGTKWDVKIRTQTVEDSGKPQASRWSDRVSITTQSLPGEIFVTVE 3043
> CE26976
Length=4280
Score = 28.1 bits (61), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 18/47 (38%), Positives = 22/47 (46%), Gaps = 1/47 (2%)
Query 30 TPPPANWPSRIRTYFVSDRWKLQAQNERLPASVRT-SFPAELFRRIE 75
T P W +IRT V D K QA S+ T S P E+F +E
Sbjct 2997 TQPGTKWDVKIRTQTVEDSGKPQASRWSDRVSITTQSLPGEIFVTVE 3043
> At1g65060
Length=561
Score = 27.3 bits (59), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 15/63 (23%), Positives = 29/63 (46%), Gaps = 4/63 (6%)
Query 4 VGSNDKKFITEHIYQIVKWKGLLDHITPPPANWPSRIRTYFVSDRWKLQAQNERLPASVR 63
V +D+ FI + + +++K+KG PPA S + + + QN+ + V
Sbjct 448 VDEDDEIFIVDRLKEVIKFKGF----QVPPAELESLLINHHSIADAAVVPQNDEVAGEVP 503
Query 64 TSF 66
+F
Sbjct 504 VAF 506
Lambda K H
0.319 0.135 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1393086308
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40