bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_1986_orf1
Length=132
Score E
Sequences producing significant alignments: (Bits) Value
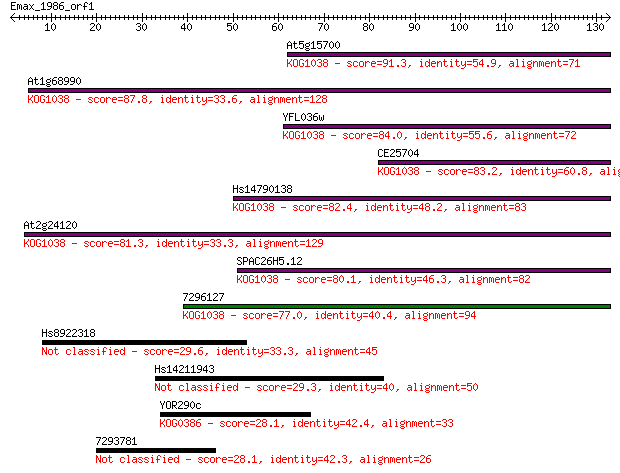
At5g15700 91.3 4e-19
At1g68990 87.8 4e-18
YFL036w 84.0 7e-17
CE25704 83.2 1e-16
Hs14790138 82.4 2e-16
At2g24120 81.3 4e-16
SPAC26H5.12 80.1 1e-15
7296127 77.0 8e-15
Hs8922318 29.6 1.4
Hs14211943 29.3 2.2
YOR290c 28.1 4.1
7293781 28.1 4.1
> At5g15700
Length=1011
Score = 91.3 bits (225), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 39/71 (54%), Positives = 48/71 (67%), Gaps = 0/71 (0%)
Query 62 SERPSFLLKLETAVNFALSSALYFPHNIDFRGRCYPLPPHLNHMGDDVSRALLRFARSRP 121
S+R LKL A A Y+PHN+DFRGR YP+PPHLNH+G D+ R +L FA RP
Sbjct 562 SQRCDTELKLSVARKMKDEEAFYYPHNMDFRGRAYPMPPHLNHLGSDLCRGVLEFAEGRP 621
Query 122 LGDSGWRWLRI 132
+G SG RWL+I
Sbjct 622 MGISGLRWLKI 632
> At1g68990
Length=976
Score = 87.8 bits (216), Expect = 4e-18, Method: Composition-based stats.
Identities = 43/128 (33%), Positives = 66/128 (51%), Gaps = 15/128 (11%)
Query 5 LGKMPSRHSMPIAEQLQQHEAQRQQQGGTAGKISSTSSSESRSLLLRHLLQQETRMKSER 64
+G + R +PI E+ ++ + ++ + K + ++Q S+R
Sbjct 485 IGGLVDREDVPIPEEPEREDQEKFKNWRWESKKA---------------IKQNNERHSQR 529
Query 65 PSFLLKLETAVNFALSSALYFPHNIDFRGRCYPLPPHLNHMGDDVSRALLRFARSRPLGD 124
LKLE A Y+PHN+DFRGR YP+ P+LNH+G D+ R +L F +PLG
Sbjct 530 CDIELKLEVARKMKDEEGFYYPHNVDFRGRAYPIHPYLNHLGSDLCRGILEFCEGKPLGK 589
Query 125 SGWRWLRI 132
SG RWL+I
Sbjct 590 SGLRWLKI 597
> YFL036w
Length=1351
Score = 84.0 bits (206), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 40/72 (55%), Positives = 48/72 (66%), Gaps = 1/72 (1%)
Query 61 KSERPSFLLKLETAVNFALSSALYFPHNIDFRGRCYPLPPHLNHMGDDVSRALLRFARSR 120
+S R KLE A F L LYFPHN+DFRGR YPL PH NH+G+D+SR LL F +
Sbjct 797 RSNRCDTNYKLEIARAF-LGEKLYFPHNLDFRGRAYPLSPHFNHLGNDMSRGLLIFWHGK 855
Query 121 PLGDSGWRWLRI 132
LG SG +WL+I
Sbjct 856 KLGPSGLKWLKI 867
> CE25704
Length=1154
Score = 83.2 bits (204), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 31/51 (60%), Positives = 44/51 (86%), Gaps = 0/51 (0%)
Query 82 ALYFPHNIDFRGRCYPLPPHLNHMGDDVSRALLRFARSRPLGDSGWRWLRI 132
L+FPHN+DFRGR YPL P+L+HMGDDV+R +L+FA+S+ LG+ G+ WL++
Sbjct 719 TLFFPHNMDFRGRVYPLSPYLSHMGDDVNRCILKFAKSQKLGEKGFDWLKL 769
> Hs14790138
Length=1230
Score = 82.4 bits (202), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 40/83 (48%), Positives = 49/83 (59%), Gaps = 1/83 (1%)
Query 50 LRHLLQQETRMKSERPSFLLKLETAVNFALSSALYFPHNIDFRGRCYPLPPHLNHMGDDV 109
L H + M S R L +L A + + PHN+DFRGR YP PPH NH+G DV
Sbjct 762 LAHCQKVAREMHSLRAEALYRLSLAQHLR-DRVFWLPHNMDFRGRTYPCPPHFNHLGSDV 820
Query 110 SRALLRFARSRPLGDSGWRWLRI 132
+RALL FA+ RPLG G WL+I
Sbjct 821 ARALLEFAQGRPLGPHGLDWLKI 843
> At2g24120
Length=993
Score = 81.3 bits (199), Expect = 4e-16, Method: Compositional matrix adjust.
Identities = 43/129 (33%), Positives = 61/129 (47%), Gaps = 15/129 (11%)
Query 4 GLGKMPSRHSMPIAEQLQQHEAQRQQQGGTAGKISSTSSSESRSLLLRHLLQQETRMKSE 63
+ + +R +PI E+ + + Q + + ++ + E SL
Sbjct 501 NIAGLVNREDVPIPEKPSSEDPEELQSWKWSARKANKINRERHSL--------------- 545
Query 64 RPSFLLKLETAVNFALSSALYFPHNIDFRGRCYPLPPHLNHMGDDVSRALLRFARSRPLG 123
R LKL A Y+PHN+DFRGR YP+ PHLNH+ D+ R L FA RPLG
Sbjct 546 RCDVELKLSVARKMKDEEGFYYPHNLDFRGRAYPMHPHLNHLSSDLCRGTLEFAEGRPLG 605
Query 124 DSGWRWLRI 132
SG WL+I
Sbjct 606 KSGLHWLKI 614
> SPAC26H5.12
Length=1120
Score = 80.1 bits (196), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 38/82 (46%), Positives = 51/82 (62%), Gaps = 1/82 (1%)
Query 51 RHLLQQETRMKSERPSFLLKLETAVNFALSSALYFPHNIDFRGRCYPLPPHLNHMGDDVS 110
+ L +T S+R F KLE A F L+ YFPH++DFRGR YPL HL+H+ +DV
Sbjct 629 KELAALKTGAHSQRCDFNYKLEIARAF-LNEKFYFPHSLDFRGRAYPLSSHLHHVSNDVC 687
Query 111 RALLRFARSRPLGDSGWRWLRI 132
R LL F+ +PLG G WL++
Sbjct 688 RGLLEFSTGKPLGPKGLNWLKV 709
> 7296127
Length=1369
Score = 77.0 bits (188), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 38/97 (39%), Positives = 60/97 (61%), Gaps = 4/97 (4%)
Query 39 STSSSESRSLLLRHLL---QQETRMKSERPSFLLKLETAVNFALSSALYFPHNIDFRGRC 95
+ +SSE R+ R L +++ M S L +L A ++ + PHN+DFRGR
Sbjct 879 NATSSEDRAKQFRDKLGHRRKQAEMYSLWCDALYRLSLAQHYR-DKVFWLPHNMDFRGRV 937
Query 96 YPLPPHLNHMGDDVSRALLRFARSRPLGDSGWRWLRI 132
YP+PPHLNH+G D++R++L F +++PLG G WL++
Sbjct 938 YPVPPHLNHLGSDLARSMLIFDQAQPLGVDGLSWLKL 974
> Hs8922318
Length=578
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 24/45 (53%), Gaps = 4/45 (8%)
Query 8 MPSRHSMPIAEQLQQHEAQRQQQGGTAGKISSTSSSESRSLLLRH 52
+P+ HS P+ E + QR G +G+I+ T + R L L+H
Sbjct 363 LPAPHSWPLLEW----QRQRHHCPGLSGRITYTPENLCRKLFLQH 403
> Hs14211943
Length=753
Score = 29.3 bits (64), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 25/50 (50%), Gaps = 3/50 (6%)
Query 33 TAGKISSTSSSESRSLLLRHLLQQETRMKSERPSFLLKLETAVNFALSSA 82
TAG I S ++S+ LLL L QET +RP +L N SSA
Sbjct 519 TAGVICSETASD---LLLHSALVQETAYIEDRPLHMLYCAAEENCLASSA 565
> YOR290c
Length=1703
Score = 28.1 bits (61), Expect = 4.1, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 34 AGKISSTSSSESRSLLLRHLLQQETRMKSERPS 66
AGK + S+SE + LLR LL E + +R S
Sbjct 1238 AGKFDNKSTSEEQEALLRSLLDAEEERRKKRES 1270
> 7293781
Length=1339
Score = 28.1 bits (61), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 11/26 (42%), Positives = 20/26 (76%), Gaps = 0/26 (0%)
Query 20 LQQHEAQRQQQGGTAGKISSTSSSES 45
+++ + QR+QQ T+GK+ +TSS+ S
Sbjct 111 VEEQDNQREQQASTSGKVKATSSTPS 136
Lambda K H
0.320 0.133 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1319765976
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40