bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
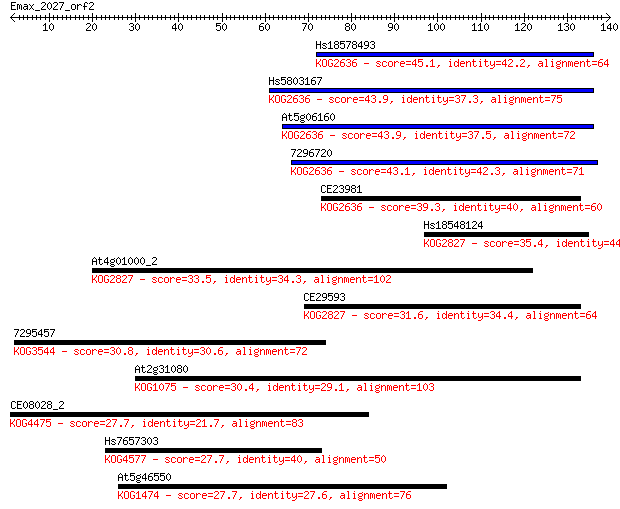
Query= Emax_2027_orf2
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
Hs18578493 45.1 5e-05
Hs5803167 43.9 9e-05
At5g06160 43.9 9e-05
7296720 43.1 1e-04
CE23981 39.3 0.002
Hs18548124 35.4 0.034
At4g01000_2 33.5 0.14
CE29593 31.6 0.54
7295457 30.8 0.75
At2g31080 30.4 1.1
CE08028_2 27.7 6.8
Hs7657303 27.7 7.1
At5g46550 27.7 7.1
> Hs18578493
Length=258
Score = 45.1 bits (105), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 27/64 (42%), Positives = 37/64 (57%), Gaps = 1/64 (1%)
Query 72 QLDVSRFNTAEELAKAVDAAVVKEKLRQLGWKCGGRPEERAARLLQLKTVDLSNVPKALL 131
LD+S F++ EELA ++ +K L LG KCG PEERA RL K L ++ +L
Sbjct 190 HLDLSAFSSWEELA-SLGLDRLKSALLALGLKCGRIPEERAQRLFSTKGKSLESLDTSLF 248
Query 132 ARPP 135
A+ P
Sbjct 249 AKNP 252
> Hs5803167
Length=501
Score = 43.9 bits (102), Expect = 9e-05, Method: Composition-based stats.
Identities = 28/75 (37%), Positives = 40/75 (53%), Gaps = 1/75 (1%)
Query 61 EAAEKVAREAQQLDVSRFNTAEELAKAVDAAVVKEKLRQLGWKCGGRPEERAARLLQLKT 120
E + + LD+S F++ EELA ++ +K L LG KCGG EERA RL K
Sbjct 232 ETSSALTHAGAHLDLSAFSSWEELA-SLGLDRLKSALLALGLKCGGTLEERAQRLFSTKG 290
Query 121 VDLSNVPKALLARPP 135
L ++ +L A+ P
Sbjct 291 KSLESLDTSLFAKNP 305
> At5g06160
Length=504
Score = 43.9 bits (102), Expect = 9e-05, Method: Composition-based stats.
Identities = 27/72 (37%), Positives = 37/72 (51%), Gaps = 1/72 (1%)
Query 64 EKVAREAQQLDVSRFNTAEELAKAVDAAVVKEKLRQLGWKCGGRPEERAARLLQLKTVDL 123
E + + +D+ ++T EEL V +KE L LG K GG P++RA RL K L
Sbjct 231 ELIPSQHTVIDLDYYSTVEELVD-VGPEKLKEALGALGLKVGGTPQQRAERLFLTKHTPL 289
Query 124 SNVPKALLARPP 135
+ K ARPP
Sbjct 290 EKLDKKHFARPP 301
> 7296720
Length=503
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 30/71 (42%), Positives = 40/71 (56%), Gaps = 3/71 (4%)
Query 66 VAREAQQLDVSRFNTAEELAKAVDAAVVKEKLRQLGWKCGGRPEERAARLLQLKTVDLSN 125
+A LD+S F++ EELA ++ +K L LG KCGG EERA RL K S
Sbjct 237 LANTGAHLDLSAFSSWEELA-SLGLDRLKSALVALGLKCGGTLEERAQRLFSTKGK--ST 293
Query 126 VPKALLARPPS 136
+ AL+A+ PS
Sbjct 294 LDPALMAKKPS 304
> CE23981
Length=500
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/60 (40%), Positives = 37/60 (61%), Gaps = 1/60 (1%)
Query 73 LDVSRFNTAEELAKAVDAAVVKEKLRQLGWKCGGRPEERAARLLQLKTVDLSNVPKALLA 132
+D+S +N+AEEL + + +K L +G KCGG +ERA RL K LS++ KA ++
Sbjct 245 VDLSPYNSAEEL-EGLGLERLKGALMAIGLKCGGTLKERADRLFATKGHKLSDLEKAAMS 303
> Hs18548124
Length=451
Score = 35.4 bits (80), Expect = 0.034, Method: Compositional matrix adjust.
Identities = 17/38 (44%), Positives = 23/38 (60%), Gaps = 0/38 (0%)
Query 97 LRQLGWKCGGRPEERAARLLQLKTVDLSNVPKALLARP 134
L LG KCGG +ERAARL ++ + + AL A+P
Sbjct 408 LMALGLKCGGTLQERAARLFSVRGLAKEQIDPALFAKP 445
> At4g01000_2
Length=317
Score = 33.5 bits (75), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 35/108 (32%), Positives = 54/108 (50%), Gaps = 12/108 (11%)
Query 20 MQQLEQMRLKASAEREKALQREAEAAAEAARAAALAEKAAAEAAEKVAREA------QQL 73
M QLE++ K+S + K L + A E +A A K A E V+ +A + L
Sbjct 203 MDQLEKVE-KSSGDAGKNL---VDVACETLITSA-AVKREGTAKETVSVDAVCCKPVEPL 257
Query 74 DVSRFNTAEELAKAVDAAVVKEKLRQLGWKCGGRPEERAARLLQLKTV 121
+ FN+ ++ + + +K +L+ G KCGG ERAARL LK+
Sbjct 258 NFDDFNSPADM-EVLGMERLKTELQSRGLKCGGTLRERAARLFLLKST 304
> CE29593
Length=500
Score = 31.6 bits (70), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 22/64 (34%), Positives = 31/64 (48%), Gaps = 1/64 (1%)
Query 69 EAQQLDVSRFNTAEELAKAVDAAVVKEKLRQLGWKCGGRPEERAARLLQLKTVDLSNVPK 128
E + + F +AE+L + + +K L G KCGG ERAARL +K PK
Sbjct 410 EYGPISLDDFTSAEDL-ELLGLEHLKSALNDRGLKCGGSLVERAARLWCVKGKQPREYPK 468
Query 129 ALLA 132
+L
Sbjct 469 NILT 472
> 7295457
Length=1180
Score = 30.8 bits (68), Expect = 0.75, Method: Composition-based stats.
Identities = 22/73 (30%), Positives = 39/73 (53%), Gaps = 1/73 (1%)
Query 2 PEKSVADSPKVDPAAAALMQQLEQMRLKASAEREKALQREAEAAAEAARAAALAEKAAAE 61
P S +V+PAAA+ + +++ A++E KA A+E +A A + AA+E
Sbjct 1093 PAASEVSKAEVEPAAASEVSKVDVEPAAAASEEVKAAVEPKPVASEEVKAPAESVPAASE 1152
Query 62 AAE-KVAREAQQL 73
+ KV EA+++
Sbjct 1153 EVKVKVEPEAEKV 1165
> At2g31080
Length=1231
Score = 30.4 bits (67), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 30/119 (25%), Positives = 54/119 (45%), Gaps = 19/119 (15%)
Query 30 ASAEREKALQREAEAAAEAARAAALAEKAAAEAAEKVAREAQQL--DVSRFNTAEELAKA 87
AS + + ++R E EA+ EK+ + V+RE +QL + S +EL K
Sbjct 554 ASVAQIRIIRRVLERFCEASGQKVSLEKSKIFFSHNVSREMEQLISEESGIGCTKELGKY 613
Query 88 VDAAVVKEKL--------------RQLGWKCGGRPEERAARLLQLKTVDLSNVPKALLA 132
+ ++++++ R GWK GR A R+ K V LS++P +++
Sbjct 614 LGMPILQKRMNKETFGEVLERVSARLAGWK--GRSLSLAGRITLTKAV-LSSIPVHVMS 669
> CE08028_2
Length=6284
Score = 27.7 bits (60), Expect = 6.8, Method: Composition-based stats.
Identities = 18/83 (21%), Positives = 37/83 (44%), Gaps = 0/83 (0%)
Query 1 NPEKSVADSPKVDPAAAALMQQLEQMRLKASAEREKALQREAEAAAEAARAAALAEKAAA 60
+PEKS + PK + ++ +K+ ++EK+ ++ E A + EK+AA
Sbjct 1403 SPEKSAEEKPKSPTKKESSPVKMADDEVKSPTKKEKSPEKVEEKPASPTKKEKTPEKSAA 1462
Query 61 EAAEKVAREAQQLDVSRFNTAEE 83
E + ++ + T +E
Sbjct 1463 EELKSPTKKEKSPSSPTKKTGDE 1485
> Hs7657303
Length=402
Score = 27.7 bits (60), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 20/55 (36%), Positives = 28/55 (50%), Gaps = 5/55 (9%)
Query 23 LEQMRLKASAEREKALQREAEAAAEAARAAALAE-----KAAAEAAEKVAREAQQ 72
+E RL A+ E A QREAEA A+ R A+ K+A + K AR ++
Sbjct 139 MEDSRLVCKADYETAKQREAEATAKRPRTTITAKQLETLKSAYNTSPKPARHVRE 193
> At5g46550
Length=506
Score = 27.7 bits (60), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 21/77 (27%), Positives = 43/77 (55%), Gaps = 1/77 (1%)
Query 26 MRLKASAEREKALQREAEAAAEA-ARAAALAEKAAAEAAEKVAREAQQLDVSRFNTAEEL 84
+R++ E+ + QRE +A EA RAA +AE+ A+ K RE+Q+L++++ +
Sbjct 382 IRIQIEKEQMERAQREEKARIEAEMRAAKVAERMRAQDELKQKRESQRLEIAKMKKGFDF 441
Query 85 AKAVDAAVVKEKLRQLG 101
+ + + K+ ++ G
Sbjct 442 ERNNHSKLKKKFVKVCG 458
Lambda K H
0.309 0.119 0.309
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1534984332
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40