bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2028_orf1
Length=139
Score E
Sequences producing significant alignments: (Bits) Value
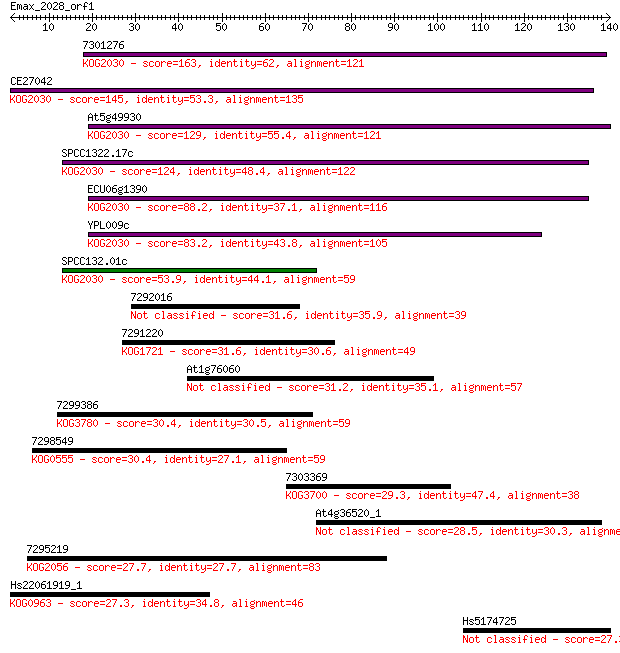
7301276 163 1e-40
CE27042 145 3e-35
At5g49930 129 1e-30
SPCC1322.17c 124 5e-29
ECU06g1390 88.2 5e-18
YPL009c 83.2 1e-16
SPCC132.01c 53.9 8e-08
7292016 31.6 0.45
7291220 31.6 0.48
At1g76060 31.2 0.64
7299386 30.4 1.2
7298549 30.4 1.2
7303369 29.3 2.2
At4g36520_1 28.5 4.5
7295219 27.7 6.8
Hs22061919_1 27.3 9.2
Hs5174725 27.3 9.9
> 7301276
Length=972
Score = 163 bits (412), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 75/121 (61%), Positives = 97/121 (80%), Gaps = 0/121 (0%)
Query 18 LVEIDISQSAYANARRMHSHRKDAAKKEQKTIDHSEKAFKNVERKVKQQLKETTIKSNIV 77
+V++D++ SA+ANARR + ++ AA+KE+KT+D S+KA K+ ERK +Q LKE SNIV
Sbjct 439 VVDVDLALSAWANARRYYDMKRSAAQKEKKTVDASQKALKSAERKTQQTLKEVRTISNIV 498
Query 78 MARKVLWFEKFFWFISSEGFLVIGGRDAQQNELIVKRYLAKDDIYVHADIHGATSIVFLP 137
ARKV WFEKF+WFISSE +LVIGGRDAQQNELIVKRY+ DIYVHA+I GA+S++
Sbjct 499 KARKVFWFEKFYWFISSENYLVIGGRDAQQNELIVKRYMRPKDIYVHAEIQGASSVIIQN 558
Query 138 P 138
P
Sbjct 559 P 559
> CE27042
Length=899
Score = 145 bits (366), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 72/135 (53%), Positives = 96/135 (71%), Gaps = 2/135 (1%)
Query 1 VMQLHDPYDPEAASKAMLVEIDISQSAYANARRMHSHRKDAAKKEQKTIDHSEKAFKNVE 60
+M L DPYD EA + + V IDIS +A NA+R +K AA+K +KT+ SEKA KN +
Sbjct 387 MMSLADPYDDEA--EVLKVPIDISLNASKNAQRHFVDKKSAAEKVKKTVASSEKAIKNAQ 444
Query 61 RKVKQQLKETTIKSNIVMARKVLWFEKFFWFISSEGFLVIGGRDAQQNELIVKRYLAKDD 120
K K L++ I + +RK +WFEKF WFISSEGF+V+ GRDAQQNEL+VK+YL +D
Sbjct 445 EKAKSTLEQVKIVVEVKKSRKSMWFEKFRWFISSEGFIVVAGRDAQQNELLVKKYLRPND 504
Query 121 IYVHADIHGATSIVF 135
IY+HAD+ GA+S+V
Sbjct 505 IYMHADVRGASSVVI 519
> At5g49930
Length=1080
Score = 129 bits (325), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 67/129 (51%), Positives = 90/129 (69%), Gaps = 8/129 (6%)
Query 19 VEIDISQSAYANARRMHSHRKDAAKKEQKTIDHSEKAFKNVERKVKQQLKETTIKSNIVM 78
VE+D+S SA+ NARR + +K K++KT+ EKAF+ E+K + QL + + + I
Sbjct 469 VEVDLSLSAHGNARRWYEMKKKQETKQEKTVSAHEKAFRAAEKKTRHQLSQEKVVATISH 528
Query 79 ARKVLWFEKFFWFISSEGFLVIGGRDAQQNELIVKRYLAKDDIYVHADIHGATSIVF--- 135
RKV WFEKF WFISSE +LVI GRDAQQNE+IVKRY++K D+YVHA++HGA+S V
Sbjct 529 MRKVHWFEKFNWFISSENYLVISGRDAQQNEMIVKRYMSKGDLYVHAELHGASSTVIKNH 588
Query 136 -----LPPL 139
+PPL
Sbjct 589 KPEQNVPPL 597
> SPCC1322.17c
Length=648
Score = 124 bits (311), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 59/124 (47%), Positives = 88/124 (70%), Gaps = 2/124 (1%)
Query 13 ASKAMLVEIDISQSAYANARRMHSHRKDAAKKEQKTIDHSEKAFKNVERKVKQQLKETTI 72
+SK + VE+D+S A+ANAR+ + R++A KE KT + + KA K+ +RK++Q LK +T
Sbjct 117 SSKFIAVELDLSLGAFANARKQYELRREALIKETKTAEAASKALKSTQRKIEQDLKRSTT 176
Query 73 KSN--IVMARKVLWFEKFFWFISSEGFLVIGGRDAQQNELIVKRYLAKDDIYVHADIHGA 130
I++ RK +FEKF WFISSEG+LV+GGRDAQQNEL+ ++Y DI+V AD+ +
Sbjct 177 ADTQRILLGRKTFFFEKFHWFISSEGYLVLGGRDAQQNELLFQKYCNTGDIFVCADLPKS 236
Query 131 TSIV 134
+ I+
Sbjct 237 SIII 240
> ECU06g1390
Length=648
Score = 88.2 bits (217), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 43/116 (37%), Positives = 71/116 (61%), Gaps = 10/116 (8%)
Query 19 VEIDISQSAYANARRMHSHRKDAAKKEQKTIDHSEKAFKNVERKVKQQLKETTIKSNIVM 78
+EID S ++N ++ K +K ++T D E+ K + K++ + +
Sbjct 363 IEIDFDSSLFSNISGLYQKSKKLEEKIRRTRDSLEEVLKRIAPKIESKK----------I 412
Query 79 ARKVLWFEKFFWFISSEGFLVIGGRDAQQNELIVKRYLAKDDIYVHADIHGATSIV 134
R WFEKF +F SS+G LVIGG++AQQNE++VK++L D+Y H+D+HG++SI+
Sbjct 413 TRAPYWFEKFHFFFSSDGVLVIGGKNAQQNEILVKKHLEPGDLYFHSDMHGSSSII 468
> YPL009c
Length=1038
Score = 83.2 bits (204), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 46/107 (42%), Positives = 68/107 (63%), Gaps = 2/107 (1%)
Query 19 VEIDISQSAYANARRMHSHRKDAAKKEQKTIDHSEKAFKNVERKVKQQLKETTIKSNIVM 78
V ID+ SAYANA + +K +A+K++K + KA KN+E K+ QQLK+ S+ V+
Sbjct 506 VTIDLGLSAYANATEYFNIKKTSAQKQKKVEKNVGKAMKNIEVKIDQQLKKKLKDSHSVL 565
Query 79 A--RKVLWFEKFFWFISSEGFLVIGGRDAQQNELIVKRYLAKDDIYV 123
R +FEK+ WFISSEGFLV+ G+ + + I +Y+ DDIY+
Sbjct 566 KKIRTPYFFEKYSWFISSEGFLVMMGKSPAETDQIYSKYIEDDDIYM 612
> SPCC132.01c
Length=549
Score = 53.9 bits (128), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 26/59 (44%), Positives = 42/59 (71%), Gaps = 0/59 (0%)
Query 13 ASKAMLVEIDISQSAYANARRMHSHRKDAAKKEQKTIDHSEKAFKNVERKVKQQLKETT 71
+SK + VE+D+S A+ANAR+ + R++A KE KT + + KA K+ +RK++Q LK +T
Sbjct 490 SSKFIAVELDLSLGAFANARKQYELRREALIKETKTAEAASKALKSTQRKIEQDLKRST 548
> 7292016
Length=469
Score = 31.6 bits (70), Expect = 0.45, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 25/39 (64%), Gaps = 0/39 (0%)
Query 29 ANARRMHSHRKDAAKKEQKTIDHSEKAFKNVERKVKQQL 67
A+ RR H RK+A+KKE + + + + VE+K +++L
Sbjct 294 ASHRRWHKPRKEASKKENRNTTNQPEKQQQVEKKSEEEL 332
> 7291220
Length=1301
Score = 31.6 bits (70), Expect = 0.48, Method: Composition-based stats.
Identities = 15/49 (30%), Positives = 28/49 (57%), Gaps = 0/49 (0%)
Query 27 AYANARRMHSHRKDAAKKEQKTIDHSEKAFKNVERKVKQQLKETTIKSN 75
+Y+ ++ H+H + KKE KT E K ++K++Q +ET K++
Sbjct 782 SYSRKQQFHAHVESHNKKEIKTFPCGECGLKFPQKKLQQHFEETGHKAD 830
> At1g76060
Length=156
Score = 31.2 bits (69), Expect = 0.64, Method: Compositional matrix adjust.
Identities = 20/57 (35%), Positives = 28/57 (49%), Gaps = 6/57 (10%)
Query 42 AKKEQKTIDHSEKAFKNVERKVKQQLKETTIKSNIVMARKVLWFEKFFWFISSEGFL 98
AKKE+ IDH E F N R +T + + + R +L +FF +I S G L
Sbjct 46 AKKEKPIIDHDEAEFLNRRRLT------STRREALSLYRDILRATRFFTWIDSRGNL 96
> 7299386
Length=457
Score = 30.4 bits (67), Expect = 1.2, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 32/64 (50%), Gaps = 5/64 (7%)
Query 12 AASKAMLVEIDISQSAYANARRMHSHRKDAAKKEQKTIDHSEKAFKNVERK-----VKQQ 66
A K M++++ S SAY +++H H + + + +D + KNV + K+Q
Sbjct 179 GAHKPMIMDVSTSYSAYVPGQKIHFHLVLDNQSDVQCMDVKVRLIKNVSYRANSSGAKEQ 238
Query 67 LKET 70
LK T
Sbjct 239 LKVT 242
> 7298549
Length=558
Score = 30.4 bits (67), Expect = 1.2, Method: Composition-based stats.
Identities = 16/64 (25%), Positives = 33/64 (51%), Gaps = 5/64 (7%)
Query 6 DPYDPEAASKAMLVEIDISQSAYANARRMHSHRKDAAKKEQ-----KTIDHSEKAFKNVE 60
+PY+P A S+ ++ + ++ NA ++ +DA K++Q + + SE V
Sbjct 63 EPYEPAAKSQLKKIQKLFVRESHKNADKLQREAEDAEKRQQNLEDARKVKISEDPSLPVA 122
Query 61 RKVK 64
RK++
Sbjct 123 RKIR 126
> 7303369
Length=697
Score = 29.3 bits (64), Expect = 2.2, Method: Composition-based stats.
Identities = 18/44 (40%), Positives = 21/44 (47%), Gaps = 6/44 (13%)
Query 65 QQLKETTIKSNIVMARKVLWFEKFF------WFISSEGFLVIGG 102
Q T I +I + R W EK F FIS + FLVIGG
Sbjct 286 HQYSVTLISPHINLFRAATWVEKPFASFILHGFISVDSFLVIGG 329
> At4g36520_1
Length=1289
Score = 28.5 bits (62), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 34/68 (50%), Gaps = 4/68 (5%)
Query 72 IKSNIVMARKVLWFEKFFWFISSEGFLVIGGRDA--QQNELIVKRYLAKDDIYVHADIHG 129
+KSNI +ARK L ++ +SS+G G D + N V+ L + ++ +AD +
Sbjct 376 VKSNIDVARKSLKDKRGSKSLSSQGSATSDGNDEWKEANNQYVE--LVRTELPRNADENS 433
Query 130 ATSIVFLP 137
V +P
Sbjct 434 GGKDVSIP 441
> 7295219
Length=810
Score = 27.7 bits (60), Expect = 6.8, Method: Composition-based stats.
Identities = 23/88 (26%), Positives = 41/88 (46%), Gaps = 8/88 (9%)
Query 5 HDPYDPEAASKAMLVEIDISQSAYANA-----RRMHSHRKDAAKKEQKTIDHSEKAFKNV 59
+DP+ P AA + + E+ + A++ +R++ H K+ + K + E K
Sbjct 32 NDPWGPSAAIMSEIAELTYNVVAFSEIMQMIWKRLNDHGKN-WRHVYKALILLEYLIKTG 90
Query 60 ERKVKQQLKETTIKSNIVMARKVLWFEK 87
KV QQ KE I R+ ++FE+
Sbjct 91 SEKVAQQCKENIFA--IQTLREFVYFEE 116
> Hs22061919_1
Length=552
Score = 27.3 bits (59), Expect = 9.2, Method: Composition-based stats.
Identities = 16/46 (34%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 1 VMQLHDPYDPEAASKAMLVEIDISQSAYANARRMHSHRKDAAKKEQ 46
+++L YD EAASKA V + ++ AN R + R+ + +EQ
Sbjct 221 LLELRRKYDEEAASKADEVGLIMTNLEKANQRAEAAQREVESLREQ 266
> Hs5174725
Length=729
Score = 27.3 bits (59), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 12/34 (35%), Positives = 18/34 (52%), Gaps = 2/34 (5%)
Query 106 QQNELIVKRYLAKDDIYVHADIHGATSIVFLPPL 139
Q+++ RY+ DI+VH D+ S PPL
Sbjct 263 QKDQYAFCRYMI--DIFVHGDLKPGQSPAIPPPL 294
Lambda K H
0.320 0.132 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1534984332
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40