bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
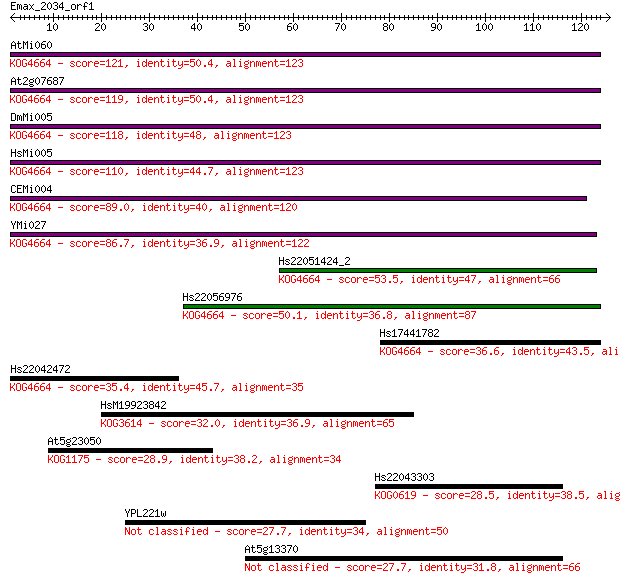
Query= Emax_2034_orf1
Length=125
Score E
Sequences producing significant alignments: (Bits) Value
AtMi060 121 3e-28
At2g07687 119 2e-27
DmMi005 118 3e-27
HsMi005 110 7e-25
CEMi004 89.0 2e-18
YMi027 86.7 9e-18
Hs22051424_2 53.5 9e-08
Hs22056976 50.1 9e-07
Hs17441782 36.6 0.011
Hs22042472 35.4 0.024
HsM19923842 32.0 0.30
At5g23050 28.9 2.5
Hs22043303 28.5 3.1
YPL221w 27.7 5.7
At5g13370 27.7 6.0
> AtMi060
Length=265
Score = 121 bits (303), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 62/123 (50%), Positives = 77/123 (62%), Gaps = 0/123 (0%)
Query 1 PPLGIASFDPKGIPFLNTVPLVSSGVSVTWTHHAIEQGDYRARIISLFLTILLGATFTSF 60
PP GI DP IPFLNT+ L+SSG +VTW HHAI G + + +L T+LL FT F
Sbjct 120 PPKGIEVLDPWEIPFLNTLILLSSGAAVTWAHHAILAGKEKRAVYALVATVLLALVFTGF 179
Query 61 QLLEYYVAGFTFSCSAYSSVYFIGTGFHGLHVLIGRVLLFICLIRFSFLVISPNHSVGFE 120
Q +EYY A FT S S Y S +F+ TGFHG HV+IG + L IC IR ++ H VGFE
Sbjct 180 QGMEYYQAPFTISDSIYGSTFFLATGFHGFHVIIGTLFLIICGIRQYLGHLTKEHHVGFE 239
Query 121 CSC 123
+
Sbjct 240 AAA 242
> At2g07687
Length=265
Score = 119 bits (297), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 62/123 (50%), Positives = 75/123 (60%), Gaps = 0/123 (0%)
Query 1 PPLGIASFDPKGIPFLNTVPLVSSGVSVTWTHHAIEQGDYRARIISLFLTILLGATFTSF 60
PP GI DP IPFLNT L SSG +VTW HHAI G + + +L T+LL FT F
Sbjct 120 PPKGIEVLDPWEIPFLNTPILPSSGAAVTWAHHAILAGKEKRAVYALVATVLLALVFTGF 179
Query 61 QLLEYYVAGFTFSCSAYSSVYFIGTGFHGLHVLIGRVLLFICLIRFSFLVISPNHSVGFE 120
Q +EYY A FT S S Y S +F+ TGFHG HV+IG + L IC IR ++ H VGFE
Sbjct 180 QGMEYYQAPFTISDSIYGSTFFLATGFHGFHVIIGTLFLIICGIRQYLGHLTKEHHVGFE 239
Query 121 CSC 123
+
Sbjct 240 AAA 242
> DmMi005
Length=262
Score = 118 bits (295), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 59/123 (47%), Positives = 77/123 (62%), Gaps = 0/123 (0%)
Query 1 PPLGIASFDPKGIPFLNTVPLVSSGVSVTWTHHAIEQGDYRARIISLFLTILLGATFTSF 60
PP+GI SF+P IP LNT L++SGV+VTW HH++ + ++ LF T+LLG FT
Sbjct 118 PPMGIISFNPFQIPLLNTAILLASGVTVTWAHHSLMENNHSQTTQGLFFTVLLGIYFTIL 177
Query 61 QLLEYYVAGFTFSCSAYSSVYFIGTGFHGLHVLIGRVLLFICLIRFSFLVISPNHSVGFE 120
Q EY A FT + S Y S +F+ TGFHG+HVLIG L +CL+R S NH GFE
Sbjct 178 QAYEYIEAPFTIADSIYGSTFFMATGFHGIHVLIGTTFLLVCLLRHLNNHFSKNHHFGFE 237
Query 121 CSC 123
+
Sbjct 238 AAA 240
> HsMi005
Length=260
Score = 110 bits (274), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 55/123 (44%), Positives = 73/123 (59%), Gaps = 0/123 (0%)
Query 1 PPLGIASFDPKGIPFLNTVPLVSSGVSVTWTHHAIEQGDYRARIISLFLTILLGATFTSF 60
PP GI +P +P LNT L++SGVS+TW HH++ + + I +L +TILLG FT
Sbjct 117 PPTGITPLNPLEVPLLNTSVLLASGVSITWAHHSLMENNRNQMIQALLITILLGLYFTLL 176
Query 61 QLLEYYVAGFTFSCSAYSSVYFIGTGFHGLHVLIGRVLLFICLIRFSFLVISPNHSVGFE 120
Q EY+ + FT S Y S +F+ TGFHGLHV+IG L IC IR + H GFE
Sbjct 177 QASEYFESPFTISDGIYGSTFFVATGFHGLHVIIGSTFLTICFIRQLMFHFTSKHHFGFE 236
Query 121 CSC 123
+
Sbjct 237 AAA 239
> CEMi004
Length=255
Score = 89.0 bits (219), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 48/120 (40%), Positives = 72/120 (60%), Gaps = 2/120 (1%)
Query 1 PPLGIASFDPKGIPFLNTVPLVSSGVSVTWTHHAIEQGDYRARIISLFLTILLGATFTSF 60
P G+ +P G+P LNT+ L+SSGV+VTW HH++ ++ S+ LT LL A FT
Sbjct 113 SPFGMHLVNPFGVPLLNTIILLSSGVTVTWAHHSLLSN--KSCTNSMILTCLLAAYFTGI 170
Query 61 QLLEYYVAGFTFSCSAYSSVYFIGTGFHGLHVLIGRVLLFICLIRFSFLVISPNHSVGFE 120
QL+EY A F+ + + S++++ TGFHG+HVL G + L +R + NH +G E
Sbjct 171 QLMEYMEASFSIADGVFGSIFYLSTGFHGIHVLCGGLFLAFNFLRLLKNHFNYNHHLGLE 230
> YMi027
Length=269
Score = 86.7 bits (213), Expect = 9e-18, Method: Compositional matrix adjust.
Identities = 45/122 (36%), Positives = 70/122 (57%), Gaps = 0/122 (0%)
Query 1 PPLGIASFDPKGIPFLNTVPLVSSGVSVTWTHHAIEQGDYRARIISLFLTILLGATFTSF 60
PP+GI + P +P LNT+ L+SSG +VT++HHA+ G+ + L +T L F +
Sbjct 125 PPVGIEAVQPTELPLLNTIILLSSGATVTYSHHALIAGNRNKALSGLLITFWLIVIFVTC 184
Query 61 QLLEYYVAGFTFSCSAYSSVYFIGTGFHGLHVLIGRVLLFICLIRFSFLVISPNHSVGFE 120
Q +EY A FT S Y SV++ GTG H LH+++ +L + R ++ H VG+E
Sbjct 185 QYIEYTNAAFTISDGVYGSVFYAGTGLHFLHMVMLAAMLGVNYWRMRNYHLTAGHHVGYE 244
Query 121 CS 122
+
Sbjct 245 TT 246
> Hs22051424_2
Length=77
Score = 53.5 bits (127), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 31/67 (46%), Positives = 37/67 (55%), Gaps = 1/67 (1%)
Query 57 FTSFQLLEYYVAGFTFSCSA-YSSVYFIGTGFHGLHVLIGRVLLFICLIRFSFLVISPNH 115
FT Q EY+ FT S Y S +F+ TGFHGLHV+IG L ICLIR + H
Sbjct 9 FTLLQASEYFETPFTISDDGIYGSTFFVATGFHGLHVIIGSTFLTICLIRQLTFHFTSKH 68
Query 116 SVGFECS 122
GFE +
Sbjct 69 HFGFEAA 75
> Hs22056976
Length=118
Score = 50.1 bits (118), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 32/90 (35%), Positives = 48/90 (53%), Gaps = 6/90 (6%)
Query 37 QGDYRARIISLFLTILLGATFTSFQLLEYYVAGFTFSCSAYSSVYFIGTGFHGLHVLIGR 96
+G + + +L +TI LG T ++ EY+ A F S Y S +F TGF GL+V+I
Sbjct 32 EGSRKQILQALAITITLGIYATLLRVSEYFEAPFPISHGLYGSTFFTATGFPGLYVIIES 91
Query 97 VLL---FICLIRFSFLVISPNHSVGFECSC 123
LL +C ++F F + NH GFE +
Sbjct 92 TLLTIRLLCQLKFHF---TSNHHFGFEATA 118
> Hs17441782
Length=102
Score = 36.6 bits (83), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 20/46 (43%), Positives = 26/46 (56%), Gaps = 1/46 (2%)
Query 78 SSVYFIGTGFHGLHVLIGRVLLFICLIRFSFLVISPNHSVGFECSC 123
S ++F GFHGLHV+I L ICL+R + NH GFE +
Sbjct 58 SEIFFFA-GFHGLHVIIRSTFLTICLLRPLKFHFTSNHHFGFEAAA 102
> Hs22042472
Length=144
Score = 35.4 bits (80), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 16/35 (45%), Positives = 23/35 (65%), Gaps = 0/35 (0%)
Query 1 PPLGIASFDPKGIPFLNTVPLVSSGVSVTWTHHAI 35
PP I+ +PFLNT L++SGVS+TW H ++
Sbjct 63 PPTSISPPSLLEVPFLNTSVLLASGVSITWAHQSL 97
> HsM19923842
Length=1095
Score = 32.0 bits (71), Expect = 0.30, Method: Composition-based stats.
Identities = 24/76 (31%), Positives = 35/76 (46%), Gaps = 11/76 (14%)
Query 20 PLVSSGVSVTWTHHAIEQGDYRARIISLFLTILLGATFTSF---------QLLEYYVAGF 70
P V + +S W R I+ LF+ L+G F SF +LL YYVA F
Sbjct 663 PGVQNFLSKQWYGEISRDTKNRKIILCLFIIPLVGCGFVSFRKKPVDKHKKLLWYYVAFF 722
Query 71 T--FSCSAYSSVYFIG 84
T F +++ V++I
Sbjct 723 TSPFVVFSWNVVFYIA 738
> At5g23050
Length=668
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 13/34 (38%), Positives = 20/34 (58%), Gaps = 2/34 (5%)
Query 9 DPKGIPFLNTVPLVSSGVSVTWTHHAIEQGDYRA 42
+PK IP+ N PL S+ + W H +++GD A
Sbjct 310 EPKAIPWTNISPLKSAADA--WCHLDVQRGDVVA 341
> Hs22043303
Length=560
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 15/39 (38%), Positives = 22/39 (56%), Gaps = 3/39 (7%)
Query 77 YSSVYFIGTGFHGLHVLIGRVLLFICLIRFSFLVISPNH 115
++S++ + G GLH L G F CL+ FL +S NH
Sbjct 279 WTSLHLLYLGNTGLHRLRGS---FRCLVNLRFLDLSQNH 314
> YPL221w
Length=793
Score = 27.7 bits (60), Expect = 5.7, Method: Composition-based stats.
Identities = 17/50 (34%), Positives = 26/50 (52%), Gaps = 3/50 (6%)
Query 25 GVSVTWTHHAIEQGDYRARIISLFLTILLGATFTSFQLLEYYVAGFTFSC 74
G S T I++ YR +I + T ++ FT F L Y++AGF +C
Sbjct 294 GNSNTLIFRGIKRMGYRMKIEN---TAIVCTGFTFFVLCGYFLAGFIMAC 340
> At5g13370
Length=573
Score = 27.7 bits (60), Expect = 6.0, Method: Composition-based stats.
Identities = 21/68 (30%), Positives = 30/68 (44%), Gaps = 2/68 (2%)
Query 50 TILLGATFTSFQLLEYYVAGFTFSCSAY-SSVYFIGTGFHGLHVLIGRVLLFI-CLIRFS 107
+I+ G LLE+Y G + S Y SS F+G F+ L I C+ F
Sbjct 277 SIITGTMAQYIPLLEFYSGGLPLTSSFYGSSECFMGVNFNPLCKPSDVSYTIIPCMGYFE 336
Query 108 FLVISPNH 115
FL + +H
Sbjct 337 FLEVEKDH 344
Lambda K H
0.331 0.146 0.468
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1183965632
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40