bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2040_orf1
Length=113
Score E
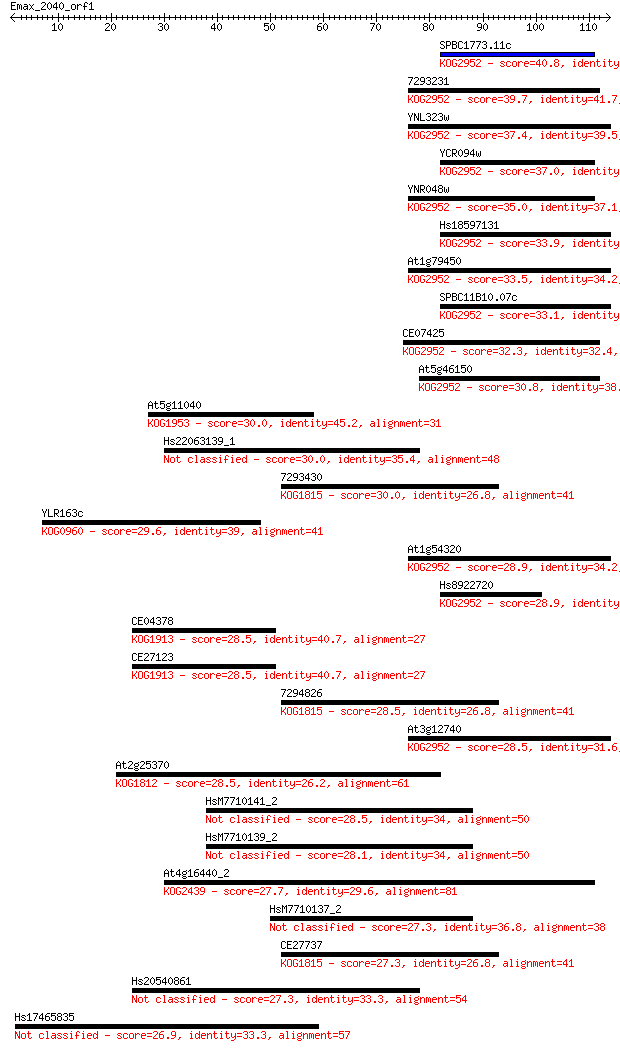
Sequences producing significant alignments: (Bits) Value
SPBC1773.11c 40.8 6e-04
7293231 39.7 0.001
YNL323w 37.4 0.007
YCR094w 37.0 0.008
YNR048w 35.0 0.030
Hs18597131 33.9 0.075
At1g79450 33.5 0.098
SPBC11B10.07c 33.1 0.13
CE07425 32.3 0.21
At5g46150 30.8 0.56
At5g11040 30.0 0.95
Hs22063139_1 30.0 1.1
7293430 30.0 1.2
YLR163c 29.6 1.3
At1g54320 28.9 2.2
Hs8922720 28.9 2.4
CE04378 28.5 2.9
CE27123 28.5 2.9
7294826 28.5 3.0
At3g12740 28.5 3.1
At2g25370 28.5 3.4
HsM7710141_2 28.5 3.6
HsM7710139_2 28.1 3.9
At4g16440_2 27.7 5.2
HsM7710137_2 27.3 6.4
CE27737 27.3 6.9
Hs20540861 27.3 7.7
Hs17465835 26.9 9.7
> SPBC1773.11c
Length=396
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 15/29 (51%), Positives = 23/29 (79%), Gaps = 0/29 (0%)
Query 82 FMQQRMSAWEPLLTPRRILFVLFGVGLIF 110
F QQR+ +W+PLLTP+ +L + F +G+IF
Sbjct 29 FRQQRIKSWQPLLTPKIVLPLFFVLGIIF 57
> 7293231
Length=357
Score = 39.7 bits (91), Expect = 0.001, Method: Composition-based stats.
Identities = 15/36 (41%), Positives = 24/36 (66%), Gaps = 0/36 (0%)
Query 76 RAALDDFMQQRMSAWEPLLTPRRILFVLFGVGLIFV 111
R + F QQR+ AW+P+LT R +L F +G++F+
Sbjct 18 RPSDSAFKQQRLPAWQPVLTARTVLPTFFVIGVLFI 53
> YNL323w
Length=414
Score = 37.4 bits (85), Expect = 0.007, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 25/38 (65%), Gaps = 0/38 (0%)
Query 76 RAALDDFMQQRMSAWEPLLTPRRILFVLFGVGLIFVVL 113
R D F QQR++A P+LTPR +L + + ++FV++
Sbjct 52 RPKEDAFTQQRLAAINPVLTPRTVLPLYLLIAVVFVIV 89
> YCR094w
Length=391
Score = 37.0 bits (84), Expect = 0.008, Method: Composition-based stats.
Identities = 14/29 (48%), Positives = 21/29 (72%), Gaps = 0/29 (0%)
Query 82 FMQQRMSAWEPLLTPRRILFVLFGVGLIF 110
F QQR+ AW+P+L+P+ +L +L V IF
Sbjct 28 FRQQRLKAWQPILSPQSVLPLLIFVACIF 56
> YNR048w
Length=393
Score = 35.0 bits (79), Expect = 0.030, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 23/35 (65%), Gaps = 0/35 (0%)
Query 76 RAALDDFMQQRMSAWEPLLTPRRILFVLFGVGLIF 110
+ A F QQR+ AW+P+L+P+ +L +L + +F
Sbjct 24 KPANTSFRQQRLKAWQPILSPQSVLPLLILMACVF 58
> Hs18597131
Length=351
Score = 33.9 bits (76), Expect = 0.075, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 82 FMQQRMSAWEPLLTPRRILFVLFGVGLIFVVL 113
F QQR+ AW+PLL+ L + F GL F+ L
Sbjct 18 FTQQRLPAWQPLLSASIALPLFFCAGLAFIGL 49
> At1g79450
Length=350
Score = 33.5 bits (75), Expect = 0.098, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 21/38 (55%), Gaps = 0/38 (0%)
Query 76 RAALDDFMQQRMSAWEPLLTPRRILFVLFGVGLIFVVL 113
R F QQ + A +P+LTPR ++ G++F+ L
Sbjct 27 RPKYSRFTQQELPACKPILTPRWVILTFLVAGVVFIPL 64
> SPBC11B10.07c
Length=371
Score = 33.1 bits (74), Expect = 0.13, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 82 FMQQRMSAWEPLLTPRRILFVLFGVGLIFVVL 113
F+QQ + AW+ + TP +L +LF +G++F L
Sbjct 23 FVQQTLPAWQFIFTPWTVLPLLFLLGIVFAPL 54
> CE07425
Length=348
Score = 32.3 bits (72), Expect = 0.21, Method: Composition-based stats.
Identities = 12/37 (32%), Positives = 22/37 (59%), Gaps = 0/37 (0%)
Query 75 SRAALDDFMQQRMSAWEPLLTPRRILFVLFGVGLIFV 111
+R QQ++ AW+P+LT ++ +F +G IF+
Sbjct 29 NRPKASALRQQKLPAWQPILTATTVIPTVFVIGAIFL 65
> At5g46150
Length=329
Score = 30.8 bits (68), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 22/34 (64%), Gaps = 0/34 (0%)
Query 78 ALDDFMQQRMSAWEPLLTPRRILFVLFGVGLIFV 111
AL F QQ++ A +P+LTP ++ V +G +F+
Sbjct 25 ALYQFKQQKLPACKPVLTPISVITVFMLMGFVFI 58
> At5g11040
Length=1186
Score = 30.0 bits (66), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 27 EEHVKQPDSPDVPESPSQELPRTRISRECSG 57
E K+ DS V +SP E P+TRI R+ S
Sbjct 914 ENSAKEDDSSPVQDSPEYEYPKTRIDRDYSA 944
> Hs22063139_1
Length=375
Score = 30.0 bits (66), Expect = 1.1, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 27/49 (55%), Gaps = 1/49 (2%)
Query 30 VKQPDSPDVPESPSQELPRTRI-SRECSGCSDGVKRSCGCGRCYCCSRA 77
+++PD DV + +++L + S+E S S G KRS GR Y + A
Sbjct 57 MEEPDRKDVAQEDAEKLGFSETDSKEASSESSGHKRSSSWGRTYSFTSA 105
> 7293430
Length=503
Score = 30.0 bits (66), Expect = 1.2, Method: Composition-based stats.
Identities = 11/41 (26%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 52 SRECSGCSDGVKRSCGCGRCYCCSRAALDDFMQQRMSAWEP 92
++EC CS +++ GC C ++ ++F + +WEP
Sbjct 288 TKECPRCSVTIEKDGGCNHMVCKNQNCKNEFCWVCLGSWEP 328
> YLR163c
Length=462
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 19/41 (46%), Gaps = 0/41 (0%)
Query 7 VAPAAGVVHHDSLCHDGQGYEEHVKQPDSPDVPESPSQELP 47
V AG V H+ L Q Y HV + +SP SP LP
Sbjct 209 VLAGAGAVDHEKLVQYAQKYFGHVPKSESPVPLGSPRGPLP 249
> At1g54320
Length=349
Score = 28.9 bits (63), Expect = 2.2, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 76 RAALDDFMQQRMSAWEPLLTPRRILFVLFGVGLIFVVL 113
R F QQ + A +P+LTP ++ V +IF+ L
Sbjct 27 RPKYSKFTQQELPACKPILTPGWVISTFLIVSVIFIPL 64
> Hs8922720
Length=361
Score = 28.9 bits (63), Expect = 2.4, Method: Composition-based stats.
Identities = 10/19 (52%), Positives = 14/19 (73%), Gaps = 0/19 (0%)
Query 82 FMQQRMSAWEPLLTPRRIL 100
F QQR+ AW+P+LT +L
Sbjct 33 FKQQRLPAWQPILTAGTVL 51
> CE04378
Length=1592
Score = 28.5 bits (62), Expect = 2.9, Method: Composition-based stats.
Identities = 11/27 (40%), Positives = 15/27 (55%), Gaps = 0/27 (0%)
Query 24 QGYEEHVKQPDSPDVPESPSQELPRTR 50
QGY+ D P+VP P Q +P T+
Sbjct 1284 QGYDLQSNAQDDPEVPREPVQHVPITQ 1310
> CE27123
Length=1596
Score = 28.5 bits (62), Expect = 2.9, Method: Composition-based stats.
Identities = 11/27 (40%), Positives = 15/27 (55%), Gaps = 0/27 (0%)
Query 24 QGYEEHVKQPDSPDVPESPSQELPRTR 50
QGY+ D P+VP P Q +P T+
Sbjct 1284 QGYDLQSNAQDDPEVPREPVQHVPITQ 1310
> 7294826
Length=511
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 11/41 (26%), Positives = 21/41 (51%), Gaps = 0/41 (0%)
Query 52 SRECSGCSDGVKRSCGCGRCYCCSRAALDDFMQQRMSAWEP 92
++EC C+ +++ GC C + + DF + +WEP
Sbjct 293 TKECPKCNVTIEKDGGCNHMVCKNPSCRYDFCWVCLGSWEP 333
> At3g12740
Length=350
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 12/38 (31%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 76 RAALDDFMQQRMSAWEPLLTPRRILFVLFGVGLIFVVL 113
R F QQ + A +P+LTP ++ + +IF+ L
Sbjct 28 RPKYSKFTQQELPACKPILTPGWVISTFLIISVIFIPL 65
> At2g25370
Length=546
Score = 28.5 bits (62), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 16/61 (26%), Positives = 26/61 (42%), Gaps = 0/61 (0%)
Query 21 HDGQGYEEHVKQPDSPDVPESPSQELPRTRISRECSGCSDGVKRSCGCGRCYCCSRAALD 80
H Y+++ K P V + + L ++ R+C C ++ S GC C A D
Sbjct 432 HTDLSYDDYKKLHPDPLVDDLKLKSLANDKMWRQCVKCRHMIELSHGCNHMTCSYDAPRD 491
Query 81 D 81
D
Sbjct 492 D 492
> HsM7710141_2
Length=385
Score = 28.5 bits (62), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 2/51 (3%)
Query 38 VPESPSQELPRTRISRECSGCSDGVKRSCGCGRC-YCCSRAALDDFMQQRM 87
V E Q R +R+C C+ ++R+ GCGRC +CC + Q+R
Sbjct 101 VDEDELQPYTNRRQNRKCGACAACLRRN-GCGRCDFCCDKPKFGGSNQKRQ 150
> HsM7710139_2
Length=430
Score = 28.1 bits (61), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 26/51 (50%), Gaps = 2/51 (3%)
Query 38 VPESPSQELPRTRISRECSGCSDGVKRSCGCGRC-YCCSRAALDDFMQQRM 87
V E Q R +R+C C+ ++R+ GCGRC +CC + Q+R
Sbjct 146 VDEDELQPYTNRRQNRKCGACAACLRRN-GCGRCDFCCDKPKFGGSNQKRQ 195
> At4g16440_2
Length=304
Score = 27.7 bits (60), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 37/88 (42%), Gaps = 14/88 (15%)
Query 30 VKQPDSPDVPESPSQELPRTRIS-RECSGCSDGVKRSCGC---GRCYCCSRAALDDFMQ- 84
VK+ D P V +P Q+L +IS ++C CS GC + +LD+F+
Sbjct 35 VKKSDRPQVVIAPKQQLEPVKISLKDCLACS-------GCITSAETVMLEKQSLDEFLSA 87
Query 85 -QRMSAWE-PLLTPRRILFVLFGVGLIF 110
+ W P L R + L+F
Sbjct 88 LSKGKTWLCPFLHSRELPLRFTMTSLLF 115
> HsM7710137_2
Length=411
Score = 27.3 bits (59), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 23/39 (58%), Gaps = 2/39 (5%)
Query 50 RISRECSGCSDGVKRSCGCGRC-YCCSRAALDDFMQQRM 87
R +R+C C+ ++R+ GCGRC +CC + Q+R
Sbjct 135 RQNRKCGACAACLRRN-GCGRCDFCCDKPKFGGSNQKRQ 172
> CE27737
Length=465
Score = 27.3 bits (59), Expect = 6.9, Method: Composition-based stats.
Identities = 11/41 (26%), Positives = 19/41 (46%), Gaps = 0/41 (0%)
Query 52 SRECSGCSDGVKRSCGCGRCYCCSRAALDDFMQQRMSAWEP 92
++EC C +++ GC C + A +F + WEP
Sbjct 279 TKECPKCMITIEKDGGCNHMTCKNTACRFEFCWMCLGPWEP 319
> Hs20540861
Length=422
Score = 27.3 bits (59), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 26/54 (48%), Gaps = 3/54 (5%)
Query 24 QGYEEHVKQPDSPDVPESPSQELPRTRISRECSGCSDGVKRSCGCGRCYCCSRA 77
+G E+ PDSP PE+ S+ PR I +E + +R+ G Y RA
Sbjct 108 KGPEDLGYSPDSPTRPETSSENTPRENIPQE---ADESGQRNRDLGLLYLHYRA 158
> Hs17465835
Length=541
Score = 26.9 bits (58), Expect = 9.7, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 30/57 (52%), Gaps = 8/57 (14%)
Query 2 MRRQKVAPAAGVVHHDSLCHDGQGYEEHVKQPDSPDVPESPSQELPRTRISRECSGC 58
+RR + G+ + S+CH QGYE+ ++P P S SQ +P ++ C GC
Sbjct 133 LRRLHLPRGEGINEYLSICHF-QGYEQGPRKP-----PRSESQPVPSKELA--CGGC 181
Lambda K H
0.325 0.139 0.459
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1185472426
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40