bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2056_orf1
Length=149
Score E
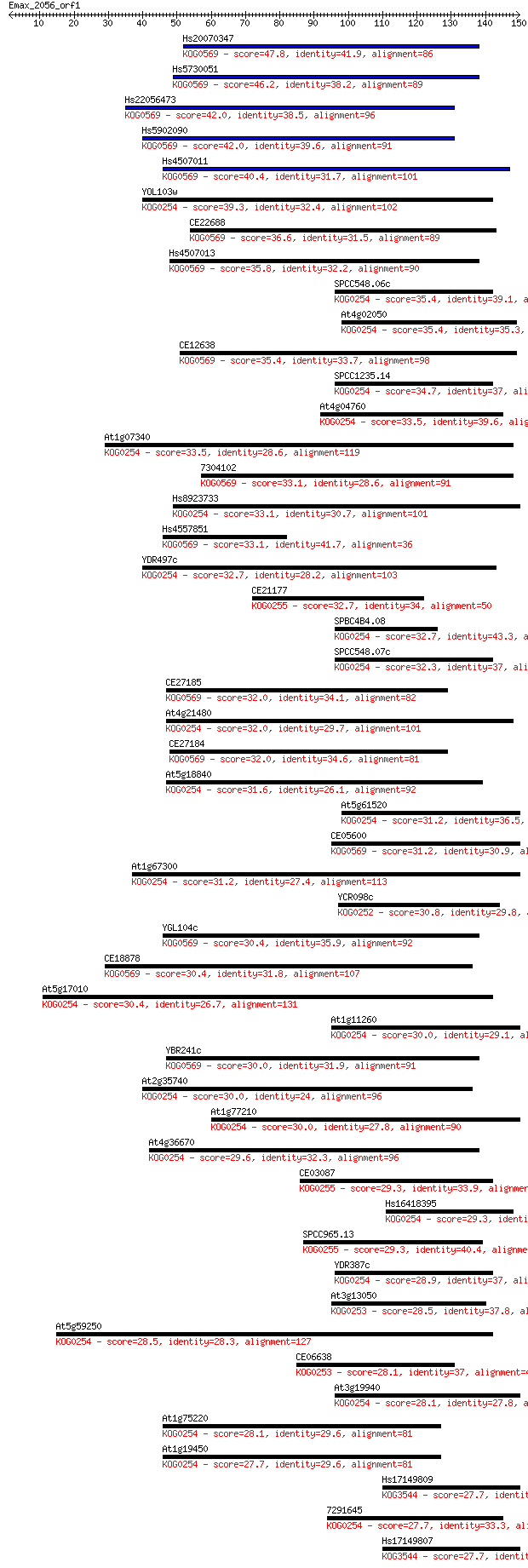
Sequences producing significant alignments: (Bits) Value
Hs20070347 47.8 7e-06
Hs5730051 46.2 2e-05
Hs22056473 42.0 5e-04
Hs5902090 42.0 5e-04
Hs4507011 40.4 0.001
YOL103w 39.3 0.003
CE22688 36.6 0.018
Hs4507013 35.8 0.027
SPCC548.06c 35.4 0.037
At4g02050 35.4 0.038
CE12638 35.4 0.044
SPCC1235.14 34.7 0.067
At4g04760 33.5 0.14
At1g07340 33.5 0.16
7304102 33.1 0.19
Hs8923733 33.1 0.20
Hs4557851 33.1 0.21
YDR497c 32.7 0.24
CE21177 32.7 0.26
SPBC4B4.08 32.7 0.28
SPCC548.07c 32.3 0.35
CE27185 32.0 0.45
At4g21480 32.0 0.45
CE27184 32.0 0.48
At5g18840 31.6 0.56
At5g61520 31.2 0.73
CE05600 31.2 0.74
At1g67300 31.2 0.85
YCR098c 30.8 1.1
YGL104c 30.4 1.3
CE18878 30.4 1.4
At5g17010 30.4 1.4
At1g11260 30.0 1.6
YBR241c 30.0 1.7
At2g35740 30.0 1.8
At1g77210 30.0 1.8
At4g36670 29.6 2.0
CE03087 29.3 3.0
Hs16418395 29.3 3.2
SPCC965.13 29.3 3.3
YDR387c 28.9 4.2
At3g13050 28.5 4.5
At5g59250 28.5 5.1
CE06638 28.1 5.8
At3g19940 28.1 6.1
At1g75220 28.1 7.0
At1g19450 27.7 7.5
Hs17149809 27.7 7.8
7291645 27.7 8.0
Hs17149807 27.7 8.1
> Hs20070347
Length=503
Score = 47.8 bits (112), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 36/94 (38%), Positives = 55/94 (58%), Gaps = 15/94 (15%)
Query 52 GSFQFGYSFSSLNTSKAIIIVDFD---WCKND----PDHVLDCSNGVLYGSLITTAVFVG 104
G+FQFGY+ S +N + + I +F W PDH++ +L SLI + +G
Sbjct 31 GTFQFGYNLSIIN-APTLHIQEFTNETWQARTGEPLPDHLV-----LLMWSLIVSLYPLG 84
Query 105 LMFGCLLAGPVT-NFGRRVSLLLTNLLFLVSSAL 137
+FG LLAGP+ GR+ SLL+ N +F+VS+A+
Sbjct 85 GLFGALLAGPLAITLGRKKSLLVNN-IFVVSAAI 117
> Hs5730051
Length=492
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 34/93 (36%), Positives = 49/93 (52%), Gaps = 6/93 (6%)
Query 49 AVLGSFQFGYSFSSLNTSKAII--IVDFDWCKNDPDHVLDCSNGVLYGSLITTAVF-VGL 105
AVLGS QFGY+ +N + +I + W + +L + L+ ++ A+F VG
Sbjct 19 AVLGSLQFGYNTGVINAPQKVIEEFYNQTWVHRYGESILPTTLTTLWS--LSVAIFSVGG 76
Query 106 MFGCLLAGPVTN-FGRRVSLLLTNLLFLVSSAL 137
M G G N FGRR S+L+ NLL VS+ L
Sbjct 77 MIGSFSVGLFVNRFGRRNSMLMMNLLAFVSAVL 109
> Hs22056473
Length=497
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 37/100 (37%), Positives = 50/100 (50%), Gaps = 6/100 (6%)
Query 35 RSCCLPAAAQFVLVAVLGSFQFGYSFSSLNTSKAIIIVDFDWCKNDPDHVLDCSNGVLYG 94
R PA + VA +GSFQFGY+ +N + II + K D + VL
Sbjct 4 RQNVTPALIFAITVATIGSFQFGYNTGVINAPETIIKEFIN--KTLTDKANAPPSEVLLT 61
Query 95 SL--ITTAVF-VGLMFGCLLAGPVTN-FGRRVSLLLTNLL 130
+L ++ A+F VG M G G N FGRR S+L+ NLL
Sbjct 62 NLWSLSVAIFSVGGMIGSFSVGLFVNRFGRRNSMLIVNLL 101
> Hs5902090
Length=496
Score = 42.0 bits (97), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 36/95 (37%), Positives = 48/95 (50%), Gaps = 6/95 (6%)
Query 40 PAAAQFVLVAVLGSFQFGYSFSSLNTSKAIIIVDFDWCKNDPDHVLDCSNGVLYGSL--I 97
PA + VA +GSFQFGY+ +N + II + D + VL SL +
Sbjct 8 PALIFAITVATIGSFQFGYNTGVINAPEKIIKEFINKTLTDKGNAPPSE--VLLTSLWSL 65
Query 98 TTAVF-VGLMFGCLLAGPVTN-FGRRVSLLLTNLL 130
+ A+F VG M G G N FGRR S+L+ NLL
Sbjct 66 SVAIFSVGGMIGSFSVGLFVNRFGRRNSMLIVNLL 100
> Hs4507011
Length=509
Score = 40.4 bits (93), Expect = 0.001, Method: Composition-based stats.
Identities = 32/107 (29%), Positives = 55/107 (51%), Gaps = 6/107 (5%)
Query 46 VLVAVLGSFQFGYSFSSLNTSKAIIIVDFD--WC-KNDPDHVLDCSNGVLYGS-LITTAV 101
V AVLGS QFGY+ +N + +I ++ W + P+ G L ++ A+
Sbjct 28 VFSAVLGSLQFGYNIGVINAPQKVIEQSYNETWLGRQGPEGPSSIPPGTLTTLWALSVAI 87
Query 102 F-VGLMFGCLLAGPVTNF-GRRVSLLLTNLLFLVSSALAAAADGVAS 146
F VG M L G ++ + GR+ ++L+ N+L ++ +L A+ AS
Sbjct 88 FSVGGMISSFLIGIISQWLGRKRAMLVNNVLAVLGGSLMGLANAAAS 134
> YOL103w
Length=612
Score = 39.3 bits (90), Expect = 0.003, Method: Composition-based stats.
Identities = 33/106 (31%), Positives = 47/106 (44%), Gaps = 17/106 (16%)
Query 40 PAAAQFVLVAVLGSFQFGYSFSSLNTSKAIIIVDFDWCKNDPDHVLDCSNGVL-YGS--L 96
P VA + F FGY ++++ I D D N VL YG L
Sbjct 110 PFIITLTFVASISGFMFGYDTGYISSALISINRDLD-------------NKVLTYGEKEL 156
Query 97 ITTAVFVGLMFGCLLAGPVTN-FGRRVSLLLTNLLFLVSSALAAAA 141
IT A +G + + AG + FGRR L+ +NL+FL+ + L A
Sbjct 157 ITAATSLGALITSVGAGTAADVFGRRPCLMFSNLMFLIGAILQITA 202
> CE22688
Length=494
Score = 36.6 bits (83), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 28/92 (30%), Positives = 47/92 (51%), Gaps = 4/92 (4%)
Query 54 FQFGYSFSSLNTSKAII--IVDFDWCKNDPDHVLDCSNGVLYGSLITTAVFVGLMFGCLL 111
FQ GY S LN A I ++ W + + D + +L+ SL+ + +FG L
Sbjct 19 FQQGYIASVLNQPYAQIEQFINSSWIERTGHPISDSTLHLLW-SLLNVCFPIATIFGQFL 77
Query 112 AGPV-TNFGRRVSLLLTNLLFLVSSALAAAAD 142
AG + + FGR+ + L+ + L++ + L AAA
Sbjct 78 AGWMCSQFGRKHTALIASFLYIPGALLCAAAK 109
> Hs4507013
Length=501
Score = 35.8 bits (81), Expect = 0.027, Method: Composition-based stats.
Identities = 29/95 (30%), Positives = 52/95 (54%), Gaps = 7/95 (7%)
Query 48 VAVLGS-FQFGYSFSSLNTSKAIIIVDF---DWCKNDPDHVLDCSNGVLYGSLITTAVFV 103
+A GS FQ+GY+ +++N S A+++ F + + + D +L+ ++ F
Sbjct 23 IAAFGSSFQYGYNVAAVN-SPALLMQQFYNETYYGRTGEFMEDFPLTLLWSVTVSMFPFG 81
Query 104 GLMFGCLLAGPVTN-FGRRVSLLLTNLLFLVSSAL 137
G + G LL GP+ N FGR+ +LL N+ +V + L
Sbjct 82 GFI-GSLLVGPLVNKFGRKGALLFNNIFSIVPAIL 115
> SPCC548.06c
Length=547
Score = 35.4 bits (80), Expect = 0.037, Method: Composition-based stats.
Identities = 18/47 (38%), Positives = 29/47 (61%), Gaps = 1/47 (2%)
Query 96 LITTAVFVGLMFGCLLAGPVTN-FGRRVSLLLTNLLFLVSSALAAAA 141
LIT V VG M GC+L+ P+ + G+RVS++ +++L+ L A
Sbjct 60 LITGMVNVGSMTGCILSSPLMDRIGKRVSIMFWTIVYLIGIILQVTA 106
> At4g02050
Length=513
Score = 35.4 bits (80), Expect = 0.038, Method: Composition-based stats.
Identities = 18/52 (34%), Positives = 32/52 (61%), Gaps = 1/52 (1%)
Query 98 TTAVFVGLMFGCLLAGPVT-NFGRRVSLLLTNLLFLVSSALAAAADGVASLF 148
T+++++ + L+A P+T N+GRR S++ + FL+ S L A A +A L
Sbjct 89 TSSLYLAGLVSTLVASPITRNYGRRASIVCGGISFLIGSGLNAGAVNLAMLL 140
> CE12638
Length=576
Score = 35.4 bits (80), Expect = 0.044, Method: Composition-based stats.
Identities = 33/103 (32%), Positives = 45/103 (43%), Gaps = 12/103 (11%)
Query 51 LGSFQFGYSFSSLNTSKAIIIVDFDWCKNDP----DHVLDCSNGVLYGSLITTAVFVGLM 106
L SFQ G+ +N +II DW K VL S+ + VG M
Sbjct 56 LASFQDGFQIGCINAPGPLII---DWIKKCHFELFGEVLSQYQADFIWSVAVSMFSVGGM 112
Query 107 FGCLLAGPVTN-FGRRVSLLLTNLLFLVSSALAAAADGVASLF 148
FG +G + + FGR+ +LL N+L L LAA + LF
Sbjct 113 FGSFCSGFLADKFGRKSTLLYNNILAL----LAAVCLSTSKLF 151
> SPCC1235.14
Length=546
Score = 34.7 bits (78), Expect = 0.067, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 27/47 (57%), Gaps = 1/47 (2%)
Query 96 LITTAVFVGLMFGCLLAGPVTN-FGRRVSLLLTNLLFLVSSALAAAA 141
LIT V VG FGC L+ P+ + G+R S++ +++L+ L A
Sbjct 60 LITGMVNVGSFFGCFLSSPLMDRIGKRTSIMFWTIVYLIGIILQVTA 106
> At4g04760
Length=457
Score = 33.5 bits (75), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 21/54 (38%), Positives = 34/54 (62%), Gaps = 5/54 (9%)
Query 92 LYGSLITTAVFVGLMFGCLLAGPVTNF-GRRVSLLLTNLLFLVSSALAAAADGV 144
L+GS++T VGL+ G L+ G +T+ GR ++ +TN+LF++ A A GV
Sbjct 51 LFGSILT----VGLILGALICGKLTDLVGRVKTIWITNILFVIGWFAIAFAKGV 100
> At1g07340
Length=468
Score = 33.5 bits (75), Expect = 0.16, Method: Composition-based stats.
Identities = 34/138 (24%), Positives = 62/138 (44%), Gaps = 37/138 (26%)
Query 29 PARFTPRS--CCLPAAAQFVLVAVLGSFQFGYSF--SSLNTSKAIIIVDF---------- 74
PA+ T + CC+ +A +G FGY S TS ++DF
Sbjct 16 PAKLTGQVFLCCV--------IAAVGGLMFGYDIGISGGVTSMDTFLLDFFPHVYEKKHR 67
Query 75 ----DWCKNDPDHVLDCSNGVLYGSLITTAVFVGLMFGCLLAGPVTN-FGRRVSLLLTNL 129
++CK D D +L L T+++++ +F ++ V+ FGR+ +++L ++
Sbjct 68 VHENNYCKFD-DQLL---------QLFTSSLYLAGIFASFISSYVSRAFGRKPTIMLASI 117
Query 130 LFLVSSALAAAADGVASL 147
FLV + L +A + L
Sbjct 118 FFLVGAILNLSAQELGML 135
> 7304102
Length=438
Score = 33.1 bits (74), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 45/96 (46%), Gaps = 8/96 (8%)
Query 57 GYSFSSLNTSKAIIIVDFDWCK----NDPDHVLDCSN-GVLYGSLITTAVFVGLMFGCLL 111
GY F LN II WC+ N+ D ++ +L+ S+++ + G+ C
Sbjct 16 GYFFGVLNAPAEIIK---KWCQDILANEYDTIVTAGQLDILWTSIVSIYLIGGICGSCFS 72
Query 112 AGPVTNFGRRVSLLLTNLLFLVSSALAAAADGVASL 147
A +GR+ L+++++LF+VS L SL
Sbjct 73 AVLCDKYGRKGCLVISSVLFVVSGILFTWCRAAKSL 108
> Hs8923733
Length=507
Score = 33.1 bits (74), Expect = 0.20, Method: Compositional matrix adjust.
Identities = 31/101 (30%), Positives = 47/101 (46%), Gaps = 8/101 (7%)
Query 49 AVLGSFQFGYSFSSLNTSKAIIIVDFDWCKNDPDHVLDCSNGVLYGSLITTAVFVGLMFG 108
AVLG+F FGY+ + TS I ++ DPD L S +GS+ T G +
Sbjct 45 AVLGNFSFGYAL--VYTSPVIPALE---RSLDPDLHLTKSQASWFGSVFTLGAAAGGLSA 99
Query 109 CLLAGPVTNFGRRVSLLLTNLLFLVSSALAAAADGVASLFL 149
+L GR++S++ + + AL A A G+ L L
Sbjct 100 MILN---DLLGRKLSIMFSAVPSAAGYALMAGAHGLWMLLL 137
> Hs4557851
Length=524
Score = 33.1 bits (74), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 46 VLVAVLGSFQFGYSFSSLNTSKAIIIVDFDWCKNDP 81
V+ AVLGSFQFGY +N + +II + P
Sbjct 14 VITAVLGSFQFGYDIGVINAPQQVIISHYRHVLGVP 49
> YDR497c
Length=584
Score = 32.7 bits (73), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 29/106 (27%), Positives = 49/106 (46%), Gaps = 15/106 (14%)
Query 40 PAAAQFVLVAVLGSFQFGYSFSSLNTSKAIIIVDFDWCKNDPDHVLDCSNGVLYGS--LI 97
P VA + F FGY + S A+I + D DH + + YG ++
Sbjct 84 PFIITLTFVASISGFMFGYDTGYI--SSALISIG-----TDLDHKV-----LTYGEKEIV 131
Query 98 TTAVFVGLMFGCLLAGPVTN-FGRRVSLLLTNLLFLVSSALAAAAD 142
T A +G + + AG + FGR+ L+ +NL+F++ + L +A
Sbjct 132 TAATSLGALITSIFAGTAADIFGRKRCLMGSNLMFVIGAILQVSAH 177
> CE21177
Length=478
Score = 32.7 bits (73), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 27/51 (52%), Gaps = 10/51 (19%)
Query 72 VDFDWCKNDPDHVLDCSNGVLYGSLITTAVFVGLMFGCLLAGPVTN-FGRR 121
++FDW C + V Y S + +VG++ G LL G +++ FGRR
Sbjct 72 IEFDWI---------CGSSVFYQSFFSQVQYVGVLIGTLLFGSLSDRFGRR 113
> SPBC4B4.08
Length=531
Score = 32.7 bits (73), Expect = 0.28, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 24/31 (77%), Gaps = 1/31 (3%)
Query 96 LITTAVFVGLMFGCLLAGPVTN-FGRRVSLL 125
L+T V VG +FGC+++ P+ + FG+R+S++
Sbjct 64 LLTGMVNVGSLFGCIISSPIADRFGKRLSII 94
> SPCC548.07c
Length=557
Score = 32.3 bits (72), Expect = 0.35, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 28/47 (59%), Gaps = 1/47 (2%)
Query 96 LITTAVFVGLMFGCLLAGPVTN-FGRRVSLLLTNLLFLVSSALAAAA 141
LIT V VG FGCL++ P T+ G+R S++ +++L+ + A
Sbjct 60 LITGMVNVGSFFGCLISSPTTDRIGKRNSIIGFCVIYLIGVIIQVTA 106
> CE27185
Length=510
Score = 32.0 bits (71), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 38/87 (43%), Gaps = 8/87 (9%)
Query 47 LVAVLGSFQFGYSFSSLNTSKAIIIVDFDWC----KNDPDHVLDCSNGVLYGSLITTAVF 102
V L SFQFGY +N +I +W K+ D L N L S+ +
Sbjct 51 FVITLASFQFGYHIGCVNAPGGLIT---EWIIGSHKDLFDKELSRENADLAWSVAVSVFA 107
Query 103 VGLMFGCLLAGPVTN-FGRRVSLLLTN 128
VG M G L +G + + GRR +L N
Sbjct 108 VGGMIGGLSSGWLADKVGRRGALFYNN 134
> At4g21480
Length=508
Score = 32.0 bits (71), Expect = 0.45, Method: Compositional matrix adjust.
Identities = 30/112 (26%), Positives = 51/112 (45%), Gaps = 14/112 (12%)
Query 47 LVAVLGSFQFGYSFSSLNTSKAIIIVD----------FDWCKNDPDHVLDCSNGVLYGSL 96
+VA +G FGY + S + +D ++ K D D C + +L
Sbjct 28 IVAAMGGLIFGYD---IGISGGVTTMDSFQQKFFPSVYEKQKKDHDSNQYCRFDSVSLTL 84
Query 97 ITTAVFVGLMFGCLLAGPVT-NFGRRVSLLLTNLLFLVSSALAAAADGVASL 147
T+++++ + L+A VT FGR++S+LL +LF + L A V L
Sbjct 85 FTSSLYLAALCSSLVASYVTRQFGRKISMLLGGVLFCAGALLNGFATAVWML 136
> CE27184
Length=492
Score = 32.0 bits (71), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 28/86 (32%), Positives = 38/86 (44%), Gaps = 8/86 (9%)
Query 48 VAVLGSFQFGYSFSSLNTSKAIIIVDFDWC----KNDPDHVLDCSNGVLYGSLITTAVFV 103
V L SFQFGY +N +I +W K+ D L N L S+ + V
Sbjct 34 VITLASFQFGYHIGCVNAPGGLIT---EWIIGSHKDLFDKELSRENADLAWSVAVSVFAV 90
Query 104 GLMFGCLLAGPVTN-FGRRVSLLLTN 128
G M G L +G + + GRR +L N
Sbjct 91 GGMIGGLSSGWLADKVGRRGALFYNN 116
> At5g18840
Length=475
Score = 31.6 bits (70), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 24/92 (26%), Positives = 45/92 (48%), Gaps = 13/92 (14%)
Query 47 LVAVLGSFQFGYSFSSLNTSKAIIIVDFDWCKNDPDHVLDCSNGVLYGSLITTAVFVGLM 106
VAV GSF+FG +++ I D + L + ++GS++T +G M
Sbjct 49 FVAVCGSFEFGSCVGYSAPTQSSIRQDLN---------LSLAEFSMFGSILT----IGAM 95
Query 107 FGCLLAGPVTNFGRRVSLLLTNLLFLVSSALA 138
G +++G +++F R + T+ F ++ LA
Sbjct 96 LGAVMSGKISDFSGRKGAMRTSACFCITGWLA 127
> At5g61520
Length=504
Score = 31.2 bits (69), Expect = 0.73, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 32/53 (60%), Gaps = 1/53 (1%)
Query 98 TTAVFVGLMFGCLLAGPVT-NFGRRVSLLLTNLLFLVSSALAAAADGVASLFL 149
T++++V + LLA VT ++GR+ S+ L + FL +AL +A VA L +
Sbjct 82 TSSLYVSGLIATLLASSVTRSWGRKPSIFLGGVSFLAGAALGGSAQNVAMLII 134
> CE05600
Length=431
Score = 31.2 bits (69), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 17/55 (30%), Positives = 29/55 (52%), Gaps = 0/55 (0%)
Query 95 SLITTAVFVGLMFGCLLAGPVTNFGRRVSLLLTNLLFLVSSALAAAADGVASLFL 149
SL ++ VG + GCL+ P++ FG + L+ N + L++ A A L+L
Sbjct 28 SLTVSSQGVGALIGCLIVSPISKFGAKDVLMRLNNVILIAGVYTGLACAFAPLYL 82
> At1g67300
Length=493
Score = 31.2 bits (69), Expect = 0.85, Method: Compositional matrix adjust.
Identities = 31/114 (27%), Positives = 48/114 (42%), Gaps = 16/114 (14%)
Query 37 CCLPAAAQFVLVAVLGSFQFGYSFSSLNTSKAIIIVDFDWCKNDPDHVLDCSNGVLYGSL 96
C LP VLVA + SF FGY +N I D L S L L
Sbjct 50 CSLP----HVLVATISSFLFGYHLGVVNEPLESISSD-----------LGFSGDTLAEGL 94
Query 97 ITTAVFVGLMFGCLLAGPVTN-FGRRVSLLLTNLLFLVSSALAAAADGVASLFL 149
+ + G G L +G V + FGRR + + L ++ + ++ ++ +A + L
Sbjct 95 VVSVCLGGAFLGSLFSGGVADGFGRRRAFQICALPMILGAFVSGVSNSLAVMLL 148
> YCR098c
Length=518
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 14/48 (29%), Positives = 27/48 (56%), Gaps = 1/48 (2%)
Query 97 ITTAVFVGLMFGCLLAGPVTN-FGRRVSLLLTNLLFLVSSALAAAADG 143
++ A VG++FG G + + R+ +L+ + ++ SAL AA+ G
Sbjct 89 VSNAALVGIIFGQFFMGIAADYYSRKSCILVATAILVIGSALCAASHG 136
> YGL104c
Length=486
Score = 30.4 bits (67), Expect = 1.3, Method: Composition-based stats.
Identities = 33/119 (27%), Positives = 45/119 (37%), Gaps = 42/119 (35%)
Query 46 VLVAVLGSFQFGYSFSSLNTSKAIIIVDFDWCKNDPDHVLDCS------NGVLYGSL--- 96
++VA +GS QFGY S LN P VL CS G Y
Sbjct 32 IIVASIGSIQFGYHLSELNA---------------PQQVLSCSEFDIPMEGYPYDRTWLG 76
Query 97 ----------------ITTAVF-VGLMFGCLLAGPVTN-FGRRVSLLLTNLLFLVSSAL 137
I T+VF +G + G A + N +GR+ S L+ L +V S +
Sbjct 77 KRGYKQCIPLNDEQIGIVTSVFCIGGILGSYFATSLANIYGRKFSSLINCTLNIVGSLI 135
> CE18878
Length=479
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 34/111 (30%), Positives = 48/111 (43%), Gaps = 11/111 (9%)
Query 29 PARFTPRSCCLPAAAQFVLVAVLGSFQFGYSFSSLNTSKAII---IVDFDWCKNDPDHVL 85
P T R A F G F FGY S++N I+ IV D +N + L
Sbjct 13 PPHLTTRLLATSVVATFA-----GGFHFGYLISAVNPLSEILQQFIV--DNLQNRYSYEL 65
Query 86 DCSNGVLYGSLITTAVFVGLMFGCLLA-GPVTNFGRRVSLLLTNLLFLVSS 135
S L S + +F+G M G ++ + + G R SLLL +L L S+
Sbjct 66 SSSQLSLIWSSLAGCLFIGAMIGAYISIFLLQSIGPRNSLLLAAILQLFSA 116
> At5g17010
Length=440
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 35/139 (25%), Positives = 58/139 (41%), Gaps = 10/139 (7%)
Query 11 EKIDATSKKEVEALGDVPPAR-------FTPRSCCLPAAAQFVLVAVLGSFQFGYSFSSL 63
E+ S E+ G++ P R P + + AA L LG +GY +
Sbjct 6 EQQQPISSVSRESSGEISPEREPLIKENHVPENYSVVAAILPFLFPALGGLLYGYEIGA- 64
Query 64 NTSKAIIIVDFDWCKNDPDHVLDCSNGVLYGSLITTAVFVGLMFGCLLAGPVTN-FGRRV 122
TS A I + V + + + +T+ G +FG ++A + + GRR
Sbjct 65 -TSCATISLQEPMTLLSYYAVPFSAVAFIKWNFMTSGSLYGALFGSIVAFTIADVIGRRK 123
Query 123 SLLLTNLLFLVSSALAAAA 141
L+L LL+LV + + A A
Sbjct 124 ELILAALLYLVGALVTALA 142
> At1g11260
Length=522
Score = 30.0 bits (66), Expect = 1.6, Method: Composition-based stats.
Identities = 16/56 (28%), Positives = 31/56 (55%), Gaps = 1/56 (1%)
Query 95 SLITTAVFVGLMFGCLLAGPVT-NFGRRVSLLLTNLLFLVSSALAAAADGVASLFL 149
++ T+++++ + L+A VT FGRR+S+L +LF + + A V L +
Sbjct 83 TMFTSSLYLAALISSLVASTVTRKFGRRLSMLFGGILFCAGALINGFAKHVWMLIV 138
> YBR241c
Length=488
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 29/104 (27%), Positives = 43/104 (41%), Gaps = 14/104 (13%)
Query 47 LVAVLGSFQFGYSFSSLNTSKAI------------IIVDFDWCKNDPDHVLDCSNGVLYG 94
+VA LGS Q+GY + LN + I D W YG
Sbjct 29 IVACLGSIQYGYHIAELNAPQEFLSCSRFEAPDENISYDDTWVGQHGLKQCIALTDSQYG 88
Query 95 SLITTAVFVGLMFGCLLAGPVTN-FGRRVSLLLTNLLFLVSSAL 137
+ IT+ +G +FG AG N +GR+ + + + +VSS L
Sbjct 89 A-ITSIFSIGGLFGSYYAGNWANRYGRKYVSMGASAMCMVSSLL 131
> At2g35740
Length=580
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 23/97 (23%), Positives = 42/97 (43%), Gaps = 11/97 (11%)
Query 40 PAAAQFVLVAVLGSFQFGYSFSSLNTSKAIIIVDFDWCKNDPDHVLDCSNGVLYGSLITT 99
P + L A +G FGY+ + + I +F + N +I +
Sbjct 24 PYIMRLALSAGIGGLLFGYNTGVIAGALLYIKEEFG----------EVDNKTWLQEIIVS 73
Query 100 AVFVGLMFGCLLAGPVTN-FGRRVSLLLTNLLFLVSS 135
G + G + G + FGRR+S+L+ ++LFL+ +
Sbjct 74 MTVAGAIVGAAIGGWYNDKFGRRMSVLIADVLFLLGA 110
> At1g77210
Length=504
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 25/91 (27%), Positives = 44/91 (48%), Gaps = 11/91 (12%)
Query 60 FSSLNTSKAIIIVDFDWCKNDPDHVLDCSNGVLYGSLITTAVFVGLMFGCLLAGPVTN-F 118
F + K + + + D+CK D + ++ + + LI+T FG A VT +
Sbjct 61 FPGIYKRKQMHLNETDYCKYDNQILTLFTSSLYFAGLIST-------FG---ASYVTRIY 110
Query 119 GRRVSLLLTNLLFLVSSALAAAADGVASLFL 149
GRR S+L+ ++ F + + AAA + L L
Sbjct 111 GRRGSILVGSVSFFLGGVINAAAKNILMLIL 141
> At4g36670
Length=493
Score = 29.6 bits (65), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 31/97 (31%), Positives = 48/97 (49%), Gaps = 14/97 (14%)
Query 42 AAQFVLVAVLGSFQFGYSFSSLNTSKAIIIVDFDWCKNDPDHVLDCSNGVLYGSLITTAV 101
A Q +VA + S FGY + S A++ ++ D ND VL G L A
Sbjct 17 ALQCAIVASIVSIIFGYDTGVM--SGAMVFIEEDLKTND------VQIEVLTGILNLCA- 67
Query 102 FVGLMFGCLLAGPVTNF-GRRVSLLLTNLLFLVSSAL 137
+ G LLAG ++ GRR +++L ++LF++ S L
Sbjct 68 ----LVGSLLAGRTSDIIGRRYTIVLASILFMLGSIL 100
> CE03087
Length=517
Score = 29.3 bits (64), Expect = 3.0, Method: Composition-based stats.
Identities = 19/57 (33%), Positives = 35/57 (61%), Gaps = 4/57 (7%)
Query 86 DCSNGVLYGSLITTAVFVGLMFGCLLAGPVTN-FGRRVSLLLTNLLFLVSSALAAAA 141
DC+ L G ++TA F+G + G + G + + +GRR ++ L++LLF + + +AA
Sbjct 109 DCTG--LNGFSLSTATFIGTLLGNIALGFLADKYGRR-TIYLSSLLFGIPCVVLSAA 162
> Hs16418395
Length=629
Score = 29.3 bits (64), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 18/39 (46%), Positives = 24/39 (61%), Gaps = 2/39 (5%)
Query 111 LAGPVTN--FGRRVSLLLTNLLFLVSSALAAAADGVASL 147
LAG N FGRR ++LL + LF SA+ AAA+ +L
Sbjct 117 LAGGALNGVFGRRAAILLASALFTAGSAVLAAANNKETL 155
> SPCC965.13
Length=537
Score = 29.3 bits (64), Expect = 3.3, Method: Composition-based stats.
Identities = 21/56 (37%), Positives = 32/56 (57%), Gaps = 6/56 (10%)
Query 87 CSNGVLYGSLITTAVFVGLMFGCLLAGPVTN-FGRR---VSLLLTNLLFLVSSALA 138
CS V Y L+ + VG FG +L GP+++ FGR+ V L+ ++F + ALA
Sbjct 122 CSIQVSY--LVQSLFIVGNAFGPMLLGPMSDIFGRKWVYVGSLILYIIFQIPQALA 175
> YDR387c
Length=555
Score = 28.9 bits (63), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 31/47 (65%), Gaps = 1/47 (2%)
Query 96 LITTAVFVGLMFGCLLAGPVTN-FGRRVSLLLTNLLFLVSSALAAAA 141
LIT++ VG FG +LA P+ + +GRR++L + +F++++ A A
Sbjct 84 LITSSTSVGSFFGSILAFPLADRYGRRITLAICCSIFILAAIGMAIA 130
> At3g13050
Length=500
Score = 28.5 bits (62), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 17/46 (36%), Positives = 28/46 (60%), Gaps = 1/46 (2%)
Query 95 SLITTAVFVGLMFGCLLAGPVTN-FGRRVSLLLTNLLFLVSSALAA 139
SLIT+ VF G++ G G V++ GRR ++T ++ V+ L+A
Sbjct 63 SLITSVVFAGMLIGAYSWGIVSDKHGRRKGFIITAVVTFVAGFLSA 108
> At5g59250
Length=579
Score = 28.5 bits (62), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 36/134 (26%), Positives = 56/134 (41%), Gaps = 24/134 (17%)
Query 15 ATSKKEVEALGDVPPARFTPRSCCLPAAAQFVLVAVLGSFQFGYSFSSLNTSKAII---- 70
A S + ++L P F+ S LP F+ A LG FGY + TS A +
Sbjct 76 ADSGEVADSLASDAPESFSWSSVILP----FIFPA-LGGLLFGYDIGA--TSGATLSLQS 128
Query 71 --IVDFDWCKNDPDHVLDCSNGVLYGSLITTAVFVGLMFGCLLAGPVTNF-GRRVSLLLT 127
+ W P + +G LYG+L+ + G V +F GRR L++
Sbjct 129 PALSGTTWFNFSPVQLGLVVSGSLYGALLGSISVYG----------VADFLGRRRELIIA 178
Query 128 NLLFLVSSALAAAA 141
+L+L+ S + A
Sbjct 179 AVLYLLGSLITGCA 192
> CE06638
Length=529
Score = 28.1 bits (61), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 26/49 (53%), Gaps = 3/49 (6%)
Query 85 LDCSNGV--LYGSLITTAVFVGLMFGCLLAGPVTN-FGRRVSLLLTNLL 130
L C G+ + +L+TT VF G+M G + + FGRR L + L+
Sbjct 110 LACEWGISSVQQALVTTCVFSGMMLSSTFWGKICDRFGRRKGLTFSTLV 158
> At3g19940
Length=514
Score = 28.1 bits (61), Expect = 6.1, Method: Composition-based stats.
Identities = 15/55 (27%), Positives = 30/55 (54%), Gaps = 1/55 (1%)
Query 96 LITTAVFVGLMFGCLLAGPVT-NFGRRVSLLLTNLLFLVSSALAAAADGVASLFL 149
L T+++++ + +A +T GR+VS+ + L FL+ + A A V+ L +
Sbjct 86 LFTSSLYLAALVASFMASVITRKHGRKVSMFIGGLAFLIGALFNAFAVNVSMLII 140
> At1g75220
Length=487
Score = 28.1 bits (61), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 39/82 (47%), Gaps = 14/82 (17%)
Query 46 VLVAVLGSFQFGYSFSSLNTSKAIIIVDFDWCKNDPDHVLDCSNGVLYGSLITTAVFVGL 105
VL+ LG QFG++ + ++A I D L S ++GSL VG
Sbjct 52 VLIVALGPIQFGFTCGYSSPTQAAITKDLG---------LTVSEYSVFGSLSN----VGA 98
Query 106 MFGCLLAGPVTNF-GRRVSLLL 126
M G + +G + + GR+ SL++
Sbjct 99 MVGAIASGQIAEYIGRKGSLMI 120
> At1g19450
Length=488
Score = 27.7 bits (60), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 24/82 (29%), Positives = 39/82 (47%), Gaps = 14/82 (17%)
Query 46 VLVAVLGSFQFGYSFSSLNTSKAIIIVDFDWCKNDPDHVLDCSNGVLYGSLITTAVFVGL 105
VL+ LG QFG++ + ++A I D L S ++GSL VG
Sbjct 53 VLIVALGPIQFGFTCGYSSPTQAAITKDLG---------LTVSEYSVFGSLSN----VGA 99
Query 106 MFGCLLAGPVTNF-GRRVSLLL 126
M G + +G + + GR+ SL++
Sbjct 100 MVGAIASGQIAEYVGRKGSLMI 121
> Hs17149809
Length=2975
Score = 27.7 bits (60), Expect = 7.8, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 22/40 (55%), Gaps = 2/40 (5%)
Query 110 LLAGPVTNFGRRVSLLLTNLLFLVSSALAAAADGVASLFL 149
LL GP N G + +L N+ LVSSA + +GV L +
Sbjct 899 LLGGPTPNTGAALEFVLRNI--LVSSAGSRITEGVPQLLI 936
> 7291645
Length=460
Score = 27.7 bits (60), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 30/52 (57%), Gaps = 1/52 (1%)
Query 94 GSLITTAVFVGLMFGCLLAGPVTNF-GRRVSLLLTNLLFLVSSALAAAADGV 144
GS I + + FG LL+GP+ ++ GRR +L+L+ + L+ + A A +
Sbjct 28 GSWIASVHSLATPFGSLLSGPLADYLGRRRTLILSVIPLLLGWSTLAIAKSI 79
> Hs17149807
Length=2976
Score = 27.7 bits (60), Expect = 8.1, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 22/40 (55%), Gaps = 2/40 (5%)
Query 110 LLAGPVTNFGRRVSLLLTNLLFLVSSALAAAADGVASLFL 149
LL GP N G + +L N+ LVSSA + +GV L +
Sbjct 900 LLGGPTPNTGAALEFVLRNI--LVSSAGSRITEGVPQLLI 937
Lambda K H
0.325 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1858150626
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40