bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2057_orf1
Length=99
Score E
Sequences producing significant alignments: (Bits) Value
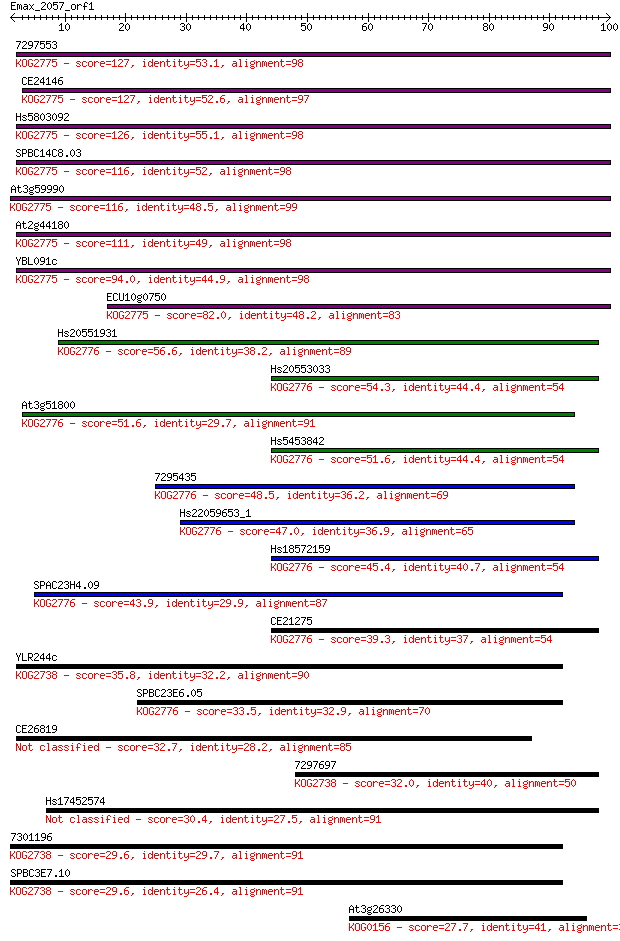
7297553 127 4e-30
CE24146 127 5e-30
Hs5803092 126 8e-30
SPBC14C8.03 116 1e-26
At3g59990 116 1e-26
At2g44180 111 3e-25
YBL091c 94.0 6e-20
ECU10g0750 82.0 2e-16
Hs20551931 56.6 1e-08
Hs20553033 54.3 6e-08
At3g51800 51.6 3e-07
Hs5453842 51.6 4e-07
7295435 48.5 3e-06
Hs22059653_1 47.0 1e-05
Hs18572159 45.4 2e-05
SPAC23H4.09 43.9 8e-05
CE21275 39.3 0.002
YLR244c 35.8 0.020
SPBC23E6.05 33.5 0.097
CE26819 32.7 0.19
7297697 32.0 0.32
Hs17452574 30.4 0.78
7301196 29.6 1.4
SPBC3E7.10 29.6 1.5
At3g26330 27.7 5.4
> 7297553
Length=448
Score = 127 bits (320), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 52/98 (53%), Positives = 71/98 (72%), Gaps = 0/98 (0%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINEC 61
LR+AAE HRQ R+Y Q ++PG+++ ++C +LE LIG G+E G FPTGCS+N C
Sbjct 139 LRQAAEAHRQTRQYMQRYIKPGMTMIQICEELENTARRLIGENGLEAGLAFPTGCSLNHC 198
Query 62 AAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
AAHYTPN G+ +L D+CK+DFG ++GRIIDCA +
Sbjct 199 AAHYTPNAGDPTVLQYDDVCKIDFGTHIKGRIIDCAFT 236
> CE24146
Length=444
Score = 127 bits (319), Expect = 5e-30, Method: Composition-based stats.
Identities = 51/97 (52%), Positives = 73/97 (75%), Gaps = 0/97 (0%)
Query 3 REAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINECA 62
R +AE HRQVR+Y +S ++PG+++ E+C +LE + LI +G+E G FPTGCS+N CA
Sbjct 134 RRSAEAHRQVRKYVKSWIKPGMTMIEICERLETTSRRLIKEQGLEAGLAFPTGCSLNHCA 193
Query 63 AHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
AHYTPN G+ +L GD+CK+D+G+ VRGR+ID A +
Sbjct 194 AHYTPNAGDTTVLQYGDVCKIDYGIHVRGRLIDSAFT 230
> Hs5803092
Length=478
Score = 126 bits (317), Expect = 8e-30, Method: Compositional matrix adjust.
Identities = 54/98 (55%), Positives = 70/98 (71%), Gaps = 0/98 (0%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINEC 61
REAAE HRQVR+Y S ++PG+++ E+C KLE + +LI G+ G FPTGCS+N C
Sbjct 169 FREAAEAHRQVRKYVMSWIKPGMTMIEICEKLEDCSRKLIKENGLNAGLAFPTGCSLNNC 228
Query 62 AAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
AAHYTPN G+ +L DICK+DFG + GRIIDCA +
Sbjct 229 AAHYTPNAGDTTVLQYDDICKIDFGTHISGRIIDCAFT 266
> SPBC14C8.03
Length=426
Score = 116 bits (290), Expect = 1e-26, Method: Composition-based stats.
Identities = 51/98 (52%), Positives = 69/98 (70%), Gaps = 0/98 (0%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINEC 61
LR AAE HRQ R+YAQS+++PG+S+ ++ +E T L+ G++ G GFPTG S+N C
Sbjct 117 LRRAAEVHRQARQYAQSVIKPGMSMMDVVNTIENTTRALVEEDGLKSGIGFPTGVSLNHC 176
Query 62 AAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
AAHYTPN G+ IL E D+ K+D GV V GRI+D A +
Sbjct 177 AAHYTPNAGDTTILKEKDVMKVDIGVHVNGRIVDSAFT 214
> At3g59990
Length=435
Score = 116 bits (290), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 48/99 (48%), Positives = 68/99 (68%), Gaps = 0/99 (0%)
Query 1 GLREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINE 60
+R AAE HRQVR+Y +S+++PG+ +T++C LE +LI G++ G FPTGCS+N
Sbjct 125 SVRRAAEVHRQVRKYVRSIVKPGMLMTDICETLENTVRKLISENGLQAGIAFPTGCSLNW 184
Query 61 CAAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
AAH+TPN G+ +L D+ KLDFG + G IIDCA +
Sbjct 185 VAAHWTPNSGDKTVLQYDDVMKLDFGTHIDGHIIDCAFT 223
> At2g44180
Length=431
Score = 111 bits (278), Expect = 3e-25, Method: Composition-based stats.
Identities = 48/98 (48%), Positives = 67/98 (68%), Gaps = 0/98 (0%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINEC 61
LR+AAE HRQVR+Y +S+L+PG+ + +LC LE +LI G++ G FPTGCS+N
Sbjct 122 LRQAAEVHRQVRKYMRSILKPGMLMIDLCETLENTVRKLISENGLQAGIAFPTGCSLNNV 181
Query 62 AAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
AAH+TPN G+ +L D+ KLDFG + G I+D A +
Sbjct 182 AAHWTPNSGDKTVLQYDDVMKLDFGTHIDGHIVDSAFT 219
> YBL091c
Length=421
Score = 94.0 bits (232), Expect = 6e-20, Method: Composition-based stats.
Identities = 44/105 (41%), Positives = 65/105 (61%), Gaps = 7/105 (6%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGI-------ERGKGFPT 54
+R+ AE HR+VRR + + PG+ L ++ +E T + GA+ + +G GFPT
Sbjct 105 VRKGAEIHRRVRRAIKDRIVPGMKLMDIADMIENTTRKYTGAENLLAMEDPKSQGIGFPT 164
Query 55 GCSINECAAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCALS 99
G S+N CAAH+TPN G+ +L D+ K+D+GVQV G IID A +
Sbjct 165 GLSLNHCAAHFTPNAGDKTVLKYEDVMKVDYGVQVNGNIIDSAFT 209
> ECU10g0750
Length=358
Score = 82.0 bits (201), Expect = 2e-16, Method: Composition-based stats.
Identities = 40/84 (47%), Positives = 54/84 (64%), Gaps = 2/84 (2%)
Query 17 QSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINECAAHYTPNPGE-DKIL 75
QS++RPG++L E+ +E T L+ + G GFP G S+N CAAHYT NPGE D +L
Sbjct 63 QSIVRPGITLLEIVRSIEDSTRTLLKGER-NNGIGFPAGMSMNSCAAHYTVNPGEQDIVL 121
Query 76 GEGDICKLDFGVQVRGRIIDCALS 99
E D+ K+DFG GRI+D A +
Sbjct 122 KEDDVLKIDFGTHSDGRIMDSAFT 145
> Hs20551931
Length=307
Score = 56.6 bits (135), Expect = 1e-08, Method: Composition-based stats.
Identities = 34/95 (35%), Positives = 50/95 (52%), Gaps = 7/95 (7%)
Query 9 HRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGA-----KGIERGKGFPTGCSINECAA 63
HR +R ++ G+ + LC K + E G K +++G FPT S+N C
Sbjct 7 HRVLRSLVEAS-SSGVLVLSLCEKSDAVIMEETGKIFKKEKEMKKGIAFPTSISVNNCVC 65
Query 64 HYTP-NPGEDKILGEGDICKLDFGVQVRGRIIDCA 97
H++P G+D IL EGD+ K+D GV V G I + A
Sbjct 66 HFSPLESGQDYILKEGDLVKIDLGVYVDGFIANVA 100
> Hs20553033
Length=210
Score = 54.3 bits (129), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 24/55 (43%), Positives = 34/55 (61%), Gaps = 1/55 (1%)
Query 44 KGIERGKGFPTGCSINECAAHYTP-NPGEDKILGEGDICKLDFGVQVRGRIIDCA 97
K +++G FPT S+N C H++P +D IL EGD+ K+D GV V G I + A
Sbjct 68 KEMKKGIAFPTSISVNNCVCHFSPLKSDQDYILKEGDLVKIDLGVHVDGFIANVA 122
> At3g51800
Length=392
Score = 51.6 bits (122), Expect = 3e-07, Method: Composition-based stats.
Identities = 27/97 (27%), Positives = 48/97 (49%), Gaps = 6/97 (6%)
Query 3 REAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGA------KGIERGKGFPTGC 56
+ AAE + + + +P + ++C K + +E + K IERG FPT
Sbjct 24 KSAAEIVNKALQVVLAECKPKAKIVDICEKGDSFIKEQTASMYKNSKKKIERGVAFPTCI 83
Query 57 SINECAAHYTPNPGEDKILGEGDICKLDFGVQVRGRI 93
S+N H++P ++ +L +GD+ K+D G + G I
Sbjct 84 SVNNTVGHFSPLASDESVLEDGDMVKIDMGCHIDGFI 120
> Hs5453842
Length=394
Score = 51.6 bits (122), Expect = 4e-07, Method: Composition-based stats.
Identities = 24/55 (43%), Positives = 34/55 (61%), Gaps = 1/55 (1%)
Query 44 KGIERGKGFPTGCSINECAAHYTP-NPGEDKILGEGDICKLDFGVQVRGRIIDCA 97
K +++G FPT S+N C H++P +D IL EGD+ K+D GV V G I + A
Sbjct 68 KEMKKGIAFPTSISVNNCVCHFSPLKSDQDYILKEGDLVKIDLGVHVDGFIANVA 122
> 7295435
Length=391
Score = 48.5 bits (114), Expect = 3e-06, Method: Composition-based stats.
Identities = 25/75 (33%), Positives = 40/75 (53%), Gaps = 6/75 (8%)
Query 25 SLTELCTKLEGKTEELIGA-----KGIERGKGFPTGCSINECAAHYTPNPGE-DKILGEG 78
S+ E+CT+ + + E G K +++G FPT S+N C H++P + D L G
Sbjct 44 SVREICTQGDNQLTEETGKVYKKEKDLKKGIAFPTCLSVNNCVCHFSPAKNDADYTLKAG 103
Query 79 DICKLDFGVQVRGRI 93
D+ K+D G + G I
Sbjct 104 DVVKIDLGAHIDGFI 118
> Hs22059653_1
Length=215
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 24/67 (35%), Positives = 39/67 (58%), Gaps = 2/67 (2%)
Query 29 LCTKLEGKTEELIG-AKGIERGKGFPTGCSINECAAHYTP-NPGEDKILGEGDICKLDFG 86
+ TK + +T ++ K +++ FPT +N C H++P +D IL EG++ K+D G
Sbjct 17 VMTKYQMETGKIFKKVKEMKKSIAFPTSILVNNCVGHFSPLKSDKDCILKEGNLVKIDLG 76
Query 87 VQVRGRI 93
VQV G I
Sbjct 77 VQVDGFI 83
> Hs18572159
Length=910
Score = 45.4 bits (106), Expect = 2e-05, Method: Composition-based stats.
Identities = 22/55 (40%), Positives = 32/55 (58%), Gaps = 1/55 (1%)
Query 44 KGIERGKGFPTGCSINECAAHYTP-NPGEDKILGEGDICKLDFGVQVRGRIIDCA 97
K +++ FPT +N C H++P +D IL EGD+ K+D GV V G I + A
Sbjct 476 KEMKKRIAFPTSILVNNCVCHFSPLKRDQDYILKEGDLVKIDLGVHVNGFITNVA 530
> SPAC23H4.09
Length=381
Score = 43.9 bits (102), Expect = 8e-05, Method: Composition-based stats.
Identities = 26/94 (27%), Positives = 44/94 (46%), Gaps = 7/94 (7%)
Query 5 AAECHRQVRRYAQSLLRPGLSLTELCTK----LEGKTEELIGAKGIERGKGFPTGCSINE 60
A E + V + L +PG + ++C + L +++ K +G FPT S N+
Sbjct 28 AGEVSQNVIKKVVELCQPGAKIYDICVRGDELLNEAIKKVYRTKDAYKGIAFPTAVSPND 87
Query 61 CAAHYTP---NPGEDKILGEGDICKLDFGVQVRG 91
AAH +P +P + L GD+ K+ G + G
Sbjct 88 MAAHLSPLKSDPEANLALKSGDVVKILLGAHIDG 121
> CE21275
Length=391
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 20/55 (36%), Positives = 26/55 (47%), Gaps = 1/55 (1%)
Query 44 KGIERGKGFPTGCSINECAAHYTPNPGEDK-ILGEGDICKLDFGVQVRGRIIDCA 97
K +G PT SI+ C HYTP E +L G + K+D G + G I A
Sbjct 79 KNFTKGIAMPTCISIDNCICHYTPLKSEAPVVLKNGQVVKVDLGTHIDGLIATAA 133
> YLR244c
Length=387
Score = 35.8 bits (81), Expect = 0.020, Method: Composition-based stats.
Identities = 29/100 (29%), Positives = 46/100 (46%), Gaps = 21/100 (21%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGK--------GFP 53
+R+A R+V A + +RPG++ EL +E++ + I+RG FP
Sbjct 138 IRKACMLGREVLDIAAAHVRPGITTDEL--------DEIVHNETIKRGAYPSPLNYYNFP 189
Query 54 TG--CSINECAAHYTPNPGEDKILGEGDICKLDFGVQVRG 91
S+NE H P + +L EGDI LD + +G
Sbjct 190 KSLCTSVNEVICHGVP---DKTVLKEGDIVNLDVSLYYQG 226
> SPBC23E6.05
Length=417
Score = 33.5 bits (75), Expect = 0.097, Method: Composition-based stats.
Identities = 23/82 (28%), Positives = 34/82 (41%), Gaps = 13/82 (15%)
Query 22 PGLSLTELCTK----LEGKTEELIGAKGIERGKGFPTGCSINECAAHYTPNPGEDKILGE 77
PG S E+ + L + ++ E+G PT +N CA +Y P P E I G
Sbjct 41 PGASTREISSYGDNLLHEYKSSIYKSQRFEKGIAEPTSICVNNCAYNYAPGP-ESVIAGN 99
Query 78 --------GDICKLDFGVQVRG 91
GD+ K+ G+ G
Sbjct 100 DNSYHLQVGDVTKISMGLHFDG 121
> CE26819
Length=498
Score = 32.7 bits (73), Expect = 0.19, Method: Composition-based stats.
Identities = 24/86 (27%), Positives = 37/86 (43%), Gaps = 3/86 (3%)
Query 2 LREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGCSINEC 61
+R A++ + R A +RPGL +L + + G + + TGC N
Sbjct 192 MRYASKIASEAHRAAMKHMRPGLYEYQLESLFRHTSYYHGGCRHLAYTCIAATGC--NGS 249
Query 62 AAHYT-PNPGEDKILGEGDICKLDFG 86
HY N DK + +GD+C D G
Sbjct 250 VLHYGHANAPNDKFIKDGDMCLFDMG 275
> 7297697
Length=307
Score = 32.0 bits (71), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 26/52 (50%), Gaps = 5/52 (9%)
Query 48 RGKGFPTG--CSINECAAHYTPNPGEDKILGEGDICKLDFGVQVRGRIIDCA 97
R GFP SIN A H P +D+ L +GDI +D V + G DC+
Sbjct 118 RYAGFPKSICTSINNIACHGIP---DDRQLADGDIINIDVTVFLNGYHGDCS 166
> Hs17452574
Length=350
Score = 30.4 bits (67), Expect = 0.78, Method: Compositional matrix adjust.
Identities = 25/93 (26%), Positives = 38/93 (40%), Gaps = 5/93 (5%)
Query 7 ECHRQVRRYAQSLLRPGLSLTELCTKLEG--KTEELIGAKGIERGKGFPTGCSINECAAH 64
EC +V + LLR G+ + L + + GA+G+E G C + + H
Sbjct 180 ECRSRVPALTRLLLRAGVIFSGLVVEHSSPPRNSPPRGAEGVESAAGSSGACGLRDLLPH 239
Query 65 YTPNPGEDKILGEGDICKLDFGVQVRGRIIDCA 97
+ P E K LG G G +RG + A
Sbjct 240 F---PHEWKALGAGQRFPAGSGDHLRGSTWNPA 269
> 7301196
Length=374
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 27/101 (26%), Positives = 40/101 (39%), Gaps = 21/101 (20%)
Query 1 GLREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERG--------KGF 52
G+R A R+ + G++ EL + L+ IER F
Sbjct 125 GMRVAGRLGRECLDEGAKAVEVGITTDEL--------DRLVHEAAIERECYPSPLNYYNF 176
Query 53 PTGC--SINECAAHYTPNPGEDKILGEGDICKLDFGVQVRG 91
P C S+NE H P + + L +GD+C +D V RG
Sbjct 177 PKSCCTSVNEVICHGIP---DQRPLQDGDLCNIDVTVYHRG 214
> SPBC3E7.10
Length=379
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 24/93 (25%), Positives = 38/93 (40%), Gaps = 5/93 (5%)
Query 1 GLREAAECHRQVRRYAQSLLRPGLSLTELCTKLEGKTEELIGAKGIERGKGFPTGC--SI 58
G+R+ R+V A + +RPG + EL + + E FP S+
Sbjct 127 GMRKVCRLGREVLDAAAAAVRPGTTTDELDSIVHNACIERDCFPSTLNYYAFPKSVCTSV 186
Query 59 NECAAHYTPNPGEDKILGEGDICKLDFGVQVRG 91
NE H P + + L +GDI +D + G
Sbjct 187 NEIICHGIP---DQRPLEDGDIVNIDVSLYHNG 216
> At3g26330
Length=500
Score = 27.7 bits (60), Expect = 5.4, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 23/41 (56%), Gaps = 2/41 (4%)
Query 57 SINECAAHYTPNPGEDKI--LGEGDICKLDFGVQVRGRIID 95
S +E AA TP +K+ L G ICK FGV +G +++
Sbjct 157 SFSESAAQKTPVNLSEKLASLTVGVICKAAFGVSFQGTVLN 197
Lambda K H
0.319 0.141 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1191270180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40