bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2094_orf1
Length=51
Score E
Sequences producing significant alignments: (Bits) Value
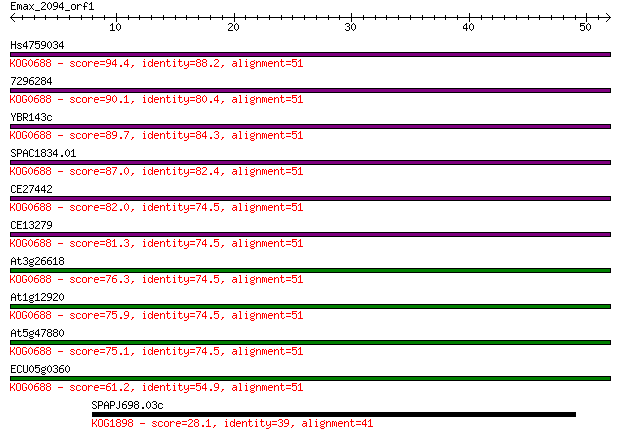
Hs4759034 94.4 5e-20
7296284 90.1 8e-19
YBR143c 89.7 1e-18
SPAC1834.01 87.0 7e-18
CE27442 82.0 2e-16
CE13279 81.3 4e-16
At3g26618 76.3 1e-14
At1g12920 75.9 2e-14
At5g47880 75.1 3e-14
ECU05g0360 61.2 4e-10
SPAPJ698.03c 28.1 3.7
> Hs4759034
Length=437
Score = 94.4 bits (233), Expect = 5e-20, Method: Composition-based stats.
Identities = 45/51 (88%), Positives = 49/51 (96%), Gaps = 0/51 (0%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGSADF 51
RGGQSALRFARLRMEKRHNY+RKVAETAVQ+FI+ DKVNV+ LVLAGSADF
Sbjct 182 RGGQSALRFARLRMEKRHNYVRKVAETAVQLFISGDKVNVAGLVLAGSADF 232
> 7296284
Length=464
Score = 90.1 bits (222), Expect = 8e-19, Method: Composition-based stats.
Identities = 41/51 (80%), Positives = 46/51 (90%), Gaps = 0/51 (0%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGSADF 51
RGGQSALRFARLRMEKRHNY+RKVAE A Q+FIT DK N++ L+LAGSADF
Sbjct 182 RGGQSALRFARLRMEKRHNYVRKVAEVATQLFITNDKPNIAGLILAGSADF 232
> YBR143c
Length=437
Score = 89.7 bits (221), Expect = 1e-18, Method: Composition-based stats.
Identities = 43/51 (84%), Positives = 45/51 (88%), Gaps = 0/51 (0%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGSADF 51
RGGQSALRFARLR EKRHNY+RKVAE AVQ FIT DKVNV L+LAGSADF
Sbjct 179 RGGQSALRFARLREEKRHNYVRKVAEVAVQNFITNDKVNVKGLILAGSADF 229
> SPAC1834.01
Length=433
Score = 87.0 bits (214), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 42/51 (82%), Positives = 45/51 (88%), Gaps = 0/51 (0%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGSADF 51
RGGQSALRFARLR EKRHNY+RKVAE AVQ FIT DK NV+ +VLAGSADF
Sbjct 179 RGGQSALRFARLRDEKRHNYVRKVAEGAVQHFITDDKPNVAGIVLAGSADF 229
> CE27442
Length=559
Score = 82.0 bits (201), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 38/51 (74%), Positives = 44/51 (86%), Gaps = 0/51 (0%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGSADF 51
RGGQSA+RFARLR EKRHNY+RKVAE +V+ FI DKV V+ L+LAGSADF
Sbjct 190 RGGQSAVRFARLRNEKRHNYVRKVAENSVEQFIKNDKVTVAGLILAGSADF 240
> CE13279
Length=443
Score = 81.3 bits (199), Expect = 4e-16, Method: Composition-based stats.
Identities = 38/51 (74%), Positives = 44/51 (86%), Gaps = 0/51 (0%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGSADF 51
RGGQSA+RFARLR EKRHNY+RKVAE +V+ FI DKV V+ L+LAGSADF
Sbjct 190 RGGQSAVRFARLRNEKRHNYVRKVAENSVEQFIKNDKVTVAGLILAGSADF 240
> At3g26618
Length=435
Score = 76.3 bits (186), Expect = 1e-14, Method: Composition-based stats.
Identities = 38/53 (71%), Positives = 42/53 (79%), Gaps = 2/53 (3%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFI--TQDKVNVSRLVLAGSADF 51
RGGQSALRFARLRMEKRHNY+RK AE A Q +I + NVS L+LAGSADF
Sbjct 180 RGGQSALRFARLRMEKRHNYVRKTAELATQFYINPATSQPNVSGLILAGSADF 232
> At1g12920
Length=434
Score = 75.9 bits (185), Expect = 2e-14, Method: Composition-based stats.
Identities = 38/53 (71%), Positives = 42/53 (79%), Gaps = 2/53 (3%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFI--TQDKVNVSRLVLAGSADF 51
RGGQSALRFARLRMEKRHNY+RK AE A Q +I + NVS L+LAGSADF
Sbjct 179 RGGQSALRFARLRMEKRHNYVRKTAELATQFYINPATSQPNVSGLILAGSADF 231
> At5g47880
Length=436
Score = 75.1 bits (183), Expect = 3e-14, Method: Composition-based stats.
Identities = 38/53 (71%), Positives = 42/53 (79%), Gaps = 2/53 (3%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFI--TQDKVNVSRLVLAGSADF 51
RGGQSALRFARLRMEKRHNY+RK AE A Q +I + NVS L+LAGSADF
Sbjct 181 RGGQSALRFARLRMEKRHNYVRKTAELATQYYINPATSQPNVSGLILAGSADF 233
> ECU05g0360
Length=386
Score = 61.2 bits (147), Expect = 4e-10, Method: Composition-based stats.
Identities = 28/51 (54%), Positives = 39/51 (76%), Gaps = 0/51 (0%)
Query 1 RGGQSALRFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGSADF 51
RGGQS++RFARLR+EKR+ Y++KV E A +F+T +NV L+LAG +D
Sbjct 175 RGGQSSVRFARLRVEKRNAYVKKVGEIAGNLFLTNCVMNVEGLILAGQSDL 225
> SPAPJ698.03c
Length=1206
Score = 28.1 bits (61), Expect = 3.7, Method: Composition-based stats.
Identities = 16/41 (39%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 8 RFARLRMEKRHNYIRKVAETAVQMFITQDKVNVSRLVLAGS 48
RF R+ +RK +AV +FIT V SR+V+A S
Sbjct 955 RFLRIYDLGNKKMLRKGELSAVPLFITHITVQASRIVVADS 995
Lambda K H
0.326 0.135 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1161614636
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40