bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2100_orf2
Length=76
Score E
Sequences producing significant alignments: (Bits) Value
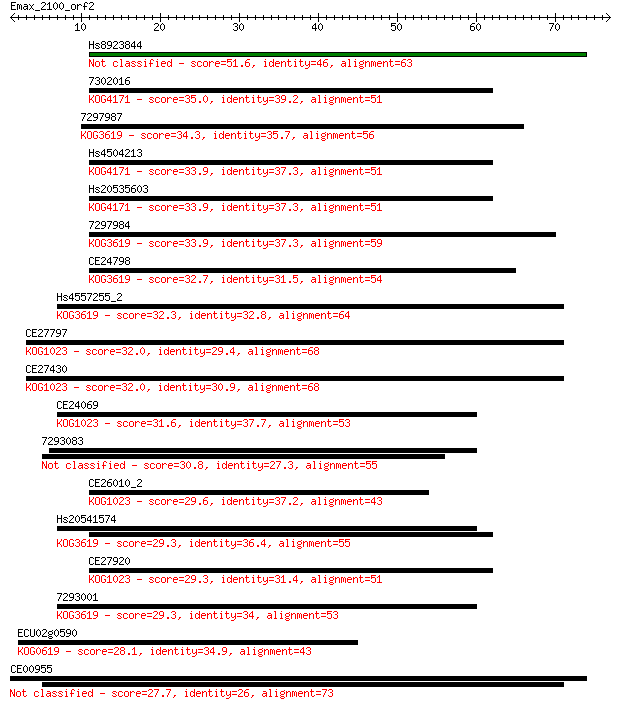
Hs8923844 51.6 3e-07
7302016 35.0 0.030
7297987 34.3 0.065
Hs4504213 33.9 0.077
Hs20535603 33.9 0.077
7297984 33.9 0.082
CE24798 32.7 0.18
Hs4557255_2 32.3 0.23
CE27797 32.0 0.28
CE27430 32.0 0.29
CE24069 31.6 0.33
7293083 30.8 0.61
CE26010_2 29.6 1.4
Hs20541574 29.3 1.7
CE27920 29.3 1.8
7293001 29.3 1.9
ECU02g0590 28.1 3.7
CE00955 27.7 4.7
> Hs8923844
Length=1610
Score = 51.6 bits (122), Expect = 3e-07, Method: Composition-based stats.
Identities = 29/64 (45%), Positives = 40/64 (62%), Gaps = 2/64 (3%)
Query 11 IGITSGRVWCGTVGNDLRKEYTALGDFVNLAARLMAKAGPREIYVDANTHRSAQ-HAMEF 69
IG+ SG V+CG VG+ +R EYT +G VNLAAR+M P + D+ T+ + A F
Sbjct 384 IGVASGIVFCGIVGHTVRHEYTVIGQKVNLAARMMMYY-PGIVTCDSVTYNGSNLPAYFF 442
Query 70 KQLP 73
K+LP
Sbjct 443 KELP 446
> 7302016
Length=787
Score = 35.0 bits (79), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 26/51 (50%), Gaps = 1/51 (1%)
Query 11 IGITSGRVWCGTVGNDLRKEYTALGDFVNLAARLMAKAGPREIYVDANTHR 61
IGI SG V G +GN + + Y G+ VNL +R P I V T+R
Sbjct 631 IGIHSGEVVTGVIGNRVPR-YCLFGNTVNLTSRTETTGVPGRINVSEETYR 680
> 7297987
Length=904
Score = 34.3 bits (77), Expect = 0.065, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 28/56 (50%), Gaps = 1/56 (1%)
Query 10 GIGITSGRVWCGTVGNDLRKEYTALGDFVNLAARLMAKAGPREIYVDANTHRSAQH 65
IGI+SG V G VG + Y G+ VN+A+R+ + P I+V T Q
Sbjct 774 AIGISSGEVMAGIVGAS-QPHYDIWGNPVNMASRMESTGLPGHIHVTEETSEILQQ 828
> Hs4504213
Length=717
Score = 33.9 bits (76), Expect = 0.077, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 29/51 (56%), Gaps = 1/51 (1%)
Query 11 IGITSGRVWCGTVGNDLRKEYTALGDFVNLAARLMAKAGPREIYVDANTHR 61
IG+ SG V+ G VG + + Y G+ V LA + + + PR+I V T+R
Sbjct 572 IGLHSGSVFAGVVGVKMPR-YCLFGNNVTLANKFESCSVPRKINVSPTTYR 621
> Hs20535603
Length=690
Score = 33.9 bits (76), Expect = 0.077, Method: Composition-based stats.
Identities = 19/51 (37%), Positives = 29/51 (56%), Gaps = 1/51 (1%)
Query 11 IGITSGRVWCGTVGNDLRKEYTALGDFVNLAARLMAKAGPREIYVDANTHR 61
IG+ SG V+ G VG + + Y G+ V LA + + + PR+I V T+R
Sbjct 575 IGLHSGSVFAGVVGVKMPR-YCLFGNNVTLANKFESCSVPRKINVSPTTYR 624
> 7297984
Length=878
Score = 33.9 bits (76), Expect = 0.082, Method: Composition-based stats.
Identities = 22/59 (37%), Positives = 31/59 (52%), Gaps = 4/59 (6%)
Query 11 IGITSGRVWCGTVGNDLRKEYTALGDFVNLAARLMAKAGPREIYVDANTHRSAQHAMEF 69
IGI+SG + G VG + Y G+ VN+A+R+ + P I V T SA+ EF
Sbjct 756 IGISSGEIMAGVVGAS-QPHYDIWGNPVNMASRMESTGLPGHIQV---TEESAKILQEF 810
> CE24798
Length=1080
Score = 32.7 bits (73), Expect = 0.18, Method: Composition-based stats.
Identities = 17/54 (31%), Positives = 29/54 (53%), Gaps = 1/54 (1%)
Query 11 IGITSGRVWCGTVGNDLRKEYTALGDFVNLAARLMAKAGPREIYVDANTHRSAQ 64
IG++ G + G +G + +Y G+ VNLA+R+ PR+I+ + R Q
Sbjct 975 IGMSVGPLVAGVIGAQ-KPQYDIWGNTVNLASRMDTHGEPRKIHATTDMGRVLQ 1027
> Hs4557255_2
Length=576
Score = 32.3 bits (72), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 33/65 (50%), Gaps = 3/65 (4%)
Query 7 LDAGIGITSGRVWCGTVGNDLRK-EYTALGDFVNLAARLMAKAGPREIYVDANTHRSAQH 65
++ +GI SG V CG +G LRK +Y V+LA R+ A P +++ T +
Sbjct 207 INMRVGIHSGNVLCGVIG--LRKWQYDVWSHDVSLANRMEAAGVPGRVHITEATLKHLDK 264
Query 66 AMEFK 70
A E +
Sbjct 265 AYEVE 269
> CE27797
Length=613
Score = 32.0 bits (71), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 29/68 (42%), Gaps = 1/68 (1%)
Query 3 KMFGLDAGIGITSGRVWCGTVGNDLRKEYTALGDFVNLAARLMAKAGPREIYVDANTHRS 62
K + L IG +G + G +G Y GD VN A+R+ + P +I T R
Sbjct 505 KNYKLRVRIGFHAGPIAAGVIGIK-NPRYCLFGDTVNFASRMQSNCPPLQIQTSEITARM 563
Query 63 AQHAMEFK 70
E+K
Sbjct 564 LLATHEYK 571
> CE27430
Length=1014
Score = 32.0 bits (71), Expect = 0.29, Method: Composition-based stats.
Identities = 21/69 (30%), Positives = 32/69 (46%), Gaps = 3/69 (4%)
Query 3 KMFGLDAGIGITSGRVWCGTVGNDLRK-EYTALGDFVNLAARLMAKAGPREIYVDANTHR 61
K + L IG +G + G +G +R Y GD VN A+R+ + P +I T R
Sbjct 916 KNYKLRIRIGFHAGPIAAGVIG--IRSPRYCLFGDTVNFASRMQSNCPPNQIQTSEITAR 973
Query 62 SAQHAMEFK 70
+ E+K
Sbjct 974 LLFDSHEYK 982
> CE24069
Length=1091
Score = 31.6 bits (70), Expect = 0.33, Method: Composition-based stats.
Identities = 20/56 (35%), Positives = 29/56 (51%), Gaps = 4/56 (7%)
Query 7 LDAGIGITSGRVWCGTVGNDLRKEYTALGDFVNLAARLMAKAGPREIYVD---ANT 59
L +GITSG V G +G+ + + GD VN+A R+ + P I + ANT
Sbjct 958 LQIKMGITSGAVAAGILGSTAPR-FCIFGDTVNMACRMASTGNPGSIQLSELTANT 1012
> 7293083
Length=1690
Score = 30.8 bits (68), Expect = 0.61, Method: Composition-based stats.
Identities = 14/54 (25%), Positives = 29/54 (53%), Gaps = 1/54 (1%)
Query 6 GLDAGIGITSGRVWCGTVGNDLRKEYTALGDFVNLAARLMAKAGPREIYVDANT 59
G+ +G+ +G V CG VG R ++ + V+LA ++ + P ++++ T
Sbjct 444 GVKMRVGVHTGTVLCGIVGTR-RVKFDVWSNDVSLANKMESSGKPEQVHISQET 496
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 18/51 (35%), Positives = 25/51 (49%), Gaps = 1/51 (1%)
Query 5 FGLDAGIGITSGRVWCGTVGNDLRKEYTALGDFVNLAARLMAKAGPREIYV 55
F L IG+ G V G +G + Y GD VN+A+R+ + P I V
Sbjct 1481 FNLILRIGMNIGDVTAGVIGTS-KLYYDIWGDAVNVASRMDSTGLPNRIQV 1530
> CE26010_2
Length=1101
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 25/43 (58%), Gaps = 1/43 (2%)
Query 11 IGITSGRVWCGTVGNDLRKEYTALGDFVNLAARLMAKAGPREI 53
+GI +G V G VG + Y GD VN+A+R+ + + P +I
Sbjct 1003 LGIHTGTVAAGVVGLTAPR-YCLFGDTVNVASRMESTSEPEKI 1044
> Hs20541574
Length=746
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 30/54 (55%), Gaps = 3/54 (5%)
Query 7 LDAGIGITSGRVWCGTVGNDLRK-EYTALGDFVNLAARLMAKAGPREIYVDANT 59
L+ +G+ +GRV CG +G LRK +Y + V LA + A P ++++ T
Sbjct 20 LNMRVGLHTGRVLCGVLG--LRKWQYDVWSNDVTLANVMEAAGLPGKVHITKTT 71
Score = 27.7 bits (60), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 17/51 (33%), Positives = 25/51 (49%), Gaps = 1/51 (1%)
Query 11 IGITSGRVWCGTVGNDLRKEYTALGDFVNLAARLMAKAGPREIYVDANTHR 61
+GI G V G +G R +Y G+ VN+A+R+ + I V HR
Sbjct 604 VGINVGPVVAGVIGAR-RPQYDIWGNTVNVASRMDSTGVQGRIQVTEEVHR 653
> CE27920
Length=1280
Score = 29.3 bits (64), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 27/51 (52%), Gaps = 1/51 (1%)
Query 11 IGITSGRVWCGTVGNDLRKEYTALGDFVNLAARLMAKAGPREIYVDANTHR 61
IG+ +G G VG + + YT GD VN A+R+ + I+ ++T +
Sbjct 1125 IGMHTGPCVAGVVGKTMPR-YTLFGDTVNTASRMESNGEALRIHCSSSTQK 1174
> 7293001
Length=2248
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 18/54 (33%), Positives = 29/54 (53%), Gaps = 3/54 (5%)
Query 7 LDAGIGITSGRVWCGTVGNDLRK-EYTALGDFVNLAARLMAKAGPREIYVDANT 59
L+ +GI +GRV CG +G LRK ++ + V LA + + P ++V T
Sbjct 365 LNMRVGIHTGRVLCGVLG--LRKWQFDVWSNDVTLANHMESGGEPGRVHVTRAT 416
> ECU02g0590
Length=785
Score = 28.1 bits (61), Expect = 3.7, Method: Composition-based stats.
Identities = 15/43 (34%), Positives = 24/43 (55%), Gaps = 4/43 (9%)
Query 2 MKMFGLDAGIGITSGRVWCGTVGNDLRKEYTALGDFVNLAARL 44
+K G+D G+GIT G + VG+ +R G +N A+R+
Sbjct 699 LKHVGIDVGVGITCGSFYTRIVGSHVR----FYGPALNKASRI 737
> CE00955
Length=1253
Score = 27.7 bits (60), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 19/79 (24%), Positives = 36/79 (45%), Gaps = 7/79 (8%)
Query 1 GMKMFGLDAG------IGITSGRVWCGTVGNDLRKEYTALGDFVNLAARLMAKAGPREIY 54
++ F +D G +GI +G+V CG VG R ++ + V LA + + ++
Sbjct 397 AIRQFDIDRGQEVNMRVGIHTGKVMCGMVGTK-RFKFDVFSNDVTLANEMESSGVAGRVH 455
Query 55 VDANTHRSAQHAMEFKQLP 73
V T + + E ++ P
Sbjct 456 VSEATAKLLKGLYEIEEGP 474
Score = 27.3 bits (59), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 18/66 (27%), Positives = 28/66 (42%), Gaps = 1/66 (1%)
Query 5 FGLDAGIGITSGRVWCGTVGNDLRKEYTALGDFVNLAARLMAKAGPREIYVDANTHRSAQ 64
F +G+ G V G +G + Y GD VN+A+R+ + I V +T
Sbjct 1150 FDFVCKLGLNIGPVTAGVIGTT-KLYYDIWGDTVNIASRMYSTGVLNRIQVSQHTREYLL 1208
Query 65 HAMEFK 70
EF+
Sbjct 1209 DRYEFE 1214
Lambda K H
0.321 0.135 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1178249128
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40