bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2173_orf1
Length=86
Score E
Sequences producing significant alignments: (Bits) Value
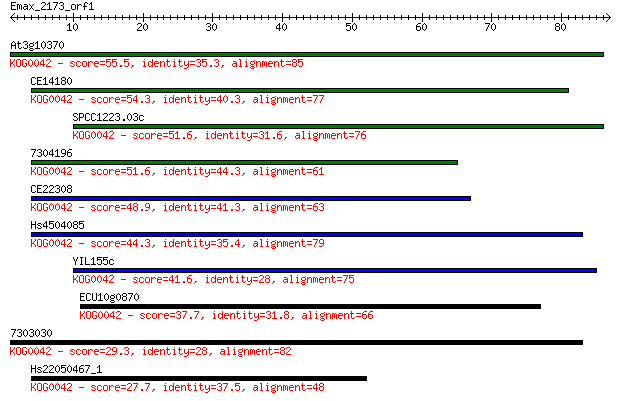
At3g10370 55.5 3e-08
CE14180 54.3 6e-08
SPCC1223.03c 51.6 3e-07
7304196 51.6 3e-07
CE22308 48.9 2e-06
Hs4504085 44.3 6e-05
YIL155c 41.6 4e-04
ECU10g0870 37.7 0.005
7303030 29.3 1.8
Hs22050467_1 27.7 5.5
> At3g10370
Length=629
Score = 55.5 bits (132), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 30/85 (35%), Positives = 44/85 (51%), Gaps = 0/85 (0%)
Query 1 LMDRLVEELPYLKAEVVFACRYEQARSVSDVIARRLPVLFLDQKLAVRVIDTMSNLMAKE 60
L RL P+L+AEV + R+E S D IARR + FLD A R + + ++A E
Sbjct 534 LGKRLAHGHPFLEAEVAYCARHEYCESAVDFIARRCRIAFLDTDAAARALQRVVEILASE 593
Query 61 LGWSASVAAKKKEEALAYAETFTVS 85
W S ++ ++A + ETF S
Sbjct 594 HKWDKSRQKQELQKAKEFLETFKSS 618
> CE14180
Length=722
Score = 54.3 bits (129), Expect = 6e-08, Method: Composition-based stats.
Identities = 31/77 (40%), Positives = 46/77 (59%), Gaps = 1/77 (1%)
Query 4 RLVEELPYLKAEVVFACRYEQARSVSDVIARRLPVLFLDQKLAVRVIDTMSNLMAKELGW 63
RL E PYL AEV +A R E A + DVIARR+ + FL+ A V+ + +M +ELGW
Sbjct 527 RLHPEFPYLDAEVRYAVR-EYACTAIDVIARRMRLAFLNTYAAHEVLPDVVRVMGQELGW 585
Query 64 SASVAAKKKEEALAYAE 80
S++ + E+A + +
Sbjct 586 SSAEQRAQLEKARTFID 602
> SPCC1223.03c
Length=649
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 24/76 (31%), Positives = 44/76 (57%), Gaps = 0/76 (0%)
Query 10 PYLKAEVVFACRYEQARSVSDVIARRLPVLFLDQKLAVRVIDTMSNLMAKELGWSASVAA 69
P+ AE+ ++ +YE R+ +D +ARR + FLD + A++ + ++++M +E GW +
Sbjct 562 PFTVAELKYSIKYEYTRTPTDFLARRTRLAFLDARQALQAVAGVTHVMKEEFGWDDATTD 621
Query 70 KKKEEALAYAETFTVS 85
K +EA Y VS
Sbjct 622 KLAQEARDYIGGMGVS 637
> 7304196
Length=713
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 27/61 (44%), Positives = 42/61 (68%), Gaps = 1/61 (1%)
Query 4 RLVEELPYLKAEVVFACRYEQARSVSDVIARRLPVLFLDQKLAVRVIDTMSNLMAKELGW 63
RL E PY++AEV R + A ++ D++ARRL V FL+ ++A ++ ++N+MAKEL W
Sbjct 507 RLHSEFPYIEAEVKQGVR-DFACTLEDMVARRLRVAFLNVQVAEEILPQVANIMAKELNW 565
Query 64 S 64
S
Sbjct 566 S 566
> CE22308
Length=673
Score = 48.9 bits (115), Expect = 2e-06, Method: Composition-based stats.
Identities = 26/63 (41%), Positives = 41/63 (65%), Gaps = 1/63 (1%)
Query 4 RLVEELPYLKAEVVFACRYEQARSVSDVIARRLPVLFLDQKLAVRVIDTMSNLMAKELGW 63
RL + P+L+AEV +A + E AR +D++ARR + LD + A +V+ + +MA+EL W
Sbjct 515 RLHPDFPFLEAEVRYAVK-EYARIPADILARRTRLSLLDARAARQVLPRIVAIMAEELKW 573
Query 64 SAS 66
S S
Sbjct 574 SPS 576
> Hs4504085
Length=727
Score = 44.3 bits (103), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 42/79 (53%), Gaps = 5/79 (6%)
Query 4 RLVEELPYLKAEVVFACRYEQARSVSDVIARRLPVLFLDQKLAVRVIDTMSNLMAKELGW 63
LV E PY++AEV + + E A + D+I+RR + FL+ + A + + LM +EL W
Sbjct 525 HLVSEFPYIEAEVKYGIK-EYACTAVDMISRRTRLAFLNVQAAEEALPRIVELMGRELNW 583
Query 64 SASVAAKKKEEALAYAETF 82
KK+E L A F
Sbjct 584 DDY----KKQEQLETARKF 598
> YIL155c
Length=649
Score = 41.6 bits (96), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 36/75 (48%), Gaps = 0/75 (0%)
Query 10 PYLKAEVVFACRYEQARSVSDVIARRLPVLFLDQKLAVRVIDTMSNLMAKELGWSASVAA 69
P+ E+ ++ +YE R+ D + RR FLD K A+ + +M E WS
Sbjct 575 PFTIGELKYSMQYEYCRTPLDFLLRRTRFAFLDAKEALNAVHATVKVMGDEFNWSEKKRQ 634
Query 70 KKKEEALAYAETFTV 84
+ E+ + + +TF V
Sbjct 635 WELEKTVNFIKTFGV 649
> ECU10g0870
Length=614
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 21/66 (31%), Positives = 34/66 (51%), Gaps = 0/66 (0%)
Query 11 YLKAEVVFACRYEQARSVSDVIARRLPVLFLDQKLAVRVIDTMSNLMAKELGWSASVAAK 70
YL EV + E A V DV+ RL + +D K A + ID + + K+ GW A +
Sbjct 507 YLIEEVEYCIDNEMAVKVCDVLCNRLMIGLMDVKEAYQCIDKVLGVFKKKHGWDADRCNR 566
Query 71 KKEEAL 76
++ +A+
Sbjct 567 EEADAI 572
> 7303030
Length=700
Score = 29.3 bits (64), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 23/89 (25%), Positives = 40/89 (44%), Gaps = 10/89 (11%)
Query 1 LMDRLVEELPYLKAEVVFACRYEQARSVSDVIARRLPVLFLDQKLAVRVIDTMSNLMAKE 60
+ +R+ E PY+ AE+ + E + D++ L D+K R I + +A E
Sbjct 517 IGNRIHPEFPYIDAEIRYGAAQEALPVIVDIMGEELG-WSKDEK--ERQIKHATEFLANE 573
Query 61 LGWSASVAAKK-------KEEALAYAETF 82
+G S + +K+ KEE Y + F
Sbjct 574 MGHSVNRTSKERIPIKLSKEEIQTYIKRF 602
> Hs22050467_1
Length=375
Score = 27.7 bits (60), Expect = 5.5, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 26/53 (49%), Gaps = 5/53 (9%)
Query 4 RLVEELPYLKAEV-----VFACRYEQARSVSDVIARRLPVLFLDQKLAVRVID 51
RLV E PY++AEV ++C S SD +R DQK + ++D
Sbjct 292 RLVSEFPYIEAEVNYGIKEYSCTTVDMISHSDRYKKRFCKSDADQKGFITIVD 344
Lambda K H
0.320 0.131 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1190857334
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40