bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2176_orf2
Length=79
Score E
Sequences producing significant alignments: (Bits) Value
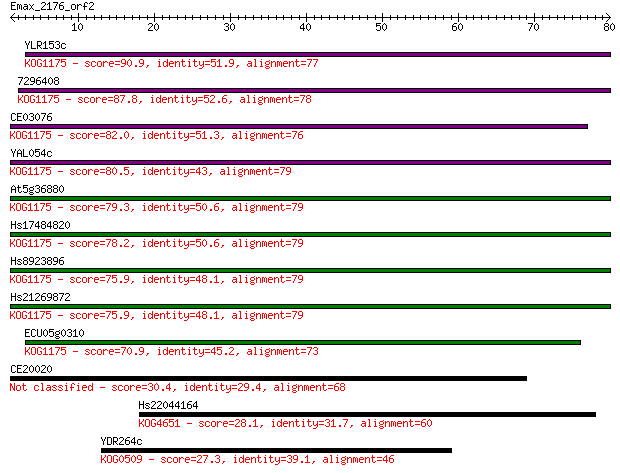
YLR153c 90.9 5e-19
7296408 87.8 5e-18
CE03076 82.0 2e-16
YAL054c 80.5 6e-16
At5g36880 79.3 1e-15
Hs17484820 78.2 3e-15
Hs8923896 75.9 2e-14
Hs21269872 75.9 2e-14
ECU05g0310 70.9 6e-13
CE20020 30.4 0.91
Hs22044164 28.1 4.4
YDR264c 27.3 6.4
> YLR153c
Length=683
Score = 90.9 bits (224), Expect = 5e-19, Method: Composition-based stats.
Identities = 40/77 (51%), Positives = 49/77 (63%), Gaps = 0/77 (0%)
Query 3 CSVVATYWQTETGGHVLAPFPGATVTKPGCAMVPFFGAEPAIVDPQSGEEKTGNNVCGVL 62
C + T WQTE+G H++AP GA TKPG A VPFFG I+DP +G E GN+V GVL
Sbjct 421 CVICDTMWQTESGSHLIAPLAGAVPTKPGSATVPFFGINACIIDPVTGVELEGNDVEGVL 480
Query 63 CIKRSWPGMCRTVHRAH 79
+K WP M R+V H
Sbjct 481 AVKSPWPSMARSVWNHH 497
> 7296408
Length=670
Score = 87.8 bits (216), Expect = 5e-18, Method: Composition-based stats.
Identities = 41/78 (52%), Positives = 51/78 (65%), Gaps = 2/78 (2%)
Query 2 QCSVVATYWQTETGGHVLAPFPGATVTKPGCAMVPFFGAEPAIVDPQSGEEKTGNNVCGV 61
QCS+V T+WQTETGGHV+ P PGAT KPG A PFFG +P ++D + G E G G
Sbjct 427 QCSIVDTFWQTETGGHVITPLPGATPMKPGSASFPFFGVKPTLLD-ECGIEIKGEGE-GY 484
Query 62 LCIKRSWPGMCRTVHRAH 79
L + WPGM RT++ H
Sbjct 485 LVFSQPWPGMMRTLYNNH 502
> CE03076
Length=680
Score = 82.0 bits (201), Expect = 2e-16, Method: Composition-based stats.
Identities = 39/77 (50%), Positives = 49/77 (63%), Gaps = 4/77 (5%)
Query 1 SQCSVVATYWQTETGGHVLAPFPGATVTKPGCAMVPFFGAEPAIVDPQSGE-EKTGNNVC 59
S S+V TYWQTETGGH++ PGAT KPG A +PFFGA P ++D + E G
Sbjct 436 SNVSIVDTYWQTETGGHMITCLPGATPMKPGAAAMPFFGASPVLLDAEGRVIEGPGE--- 492
Query 60 GVLCIKRSWPGMCRTVH 76
G LC R+WPGM R ++
Sbjct 493 GSLCFDRAWPGMMRGIY 509
> YAL054c
Length=713
Score = 80.5 bits (197), Expect = 6e-16, Method: Composition-based stats.
Identities = 34/80 (42%), Positives = 50/80 (62%), Gaps = 1/80 (1%)
Query 1 SQCSVVATYWQTETGGHVLAPFPGA-TVTKPGCAMVPFFGAEPAIVDPQSGEEKTGNNVC 59
++ +V TYWQTE+G H++ P G T KPG A PFFG + ++DP +GEE ++
Sbjct 461 NEIPIVDTYWQTESGSHLVTPLAGGVTPMKPGSASFPFFGIDAVVLDPNTGEELNTSHAE 520
Query 60 GVLCIKRSWPGMCRTVHRAH 79
GVL +K +WP RT+ + H
Sbjct 521 GVLAVKAAWPSFARTIWKNH 540
> At5g36880
Length=693
Score = 79.3 bits (194), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 40/79 (50%), Positives = 49/79 (62%), Gaps = 2/79 (2%)
Query 1 SQCSVVATYWQTETGGHVLAPFPGATVTKPGCAMVPFFGAEPAIVDPQSGEEKTGNNVCG 60
S+C + T+WQTETGG ++ P PGA KPG A PFFG +P IVD + G E G G
Sbjct 451 SRCPISDTWWQTETGGFMITPLPGAWPQKPGSATFPFFGVQPVIVD-EKGNEIEG-ECSG 508
Query 61 VLCIKRSWPGMCRTVHRAH 79
LC+K SWPG RT+ H
Sbjct 509 YLCVKGSWPGAFRTLFGDH 527
> Hs17484820
Length=430
Score = 78.2 bits (191), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 40/82 (48%), Positives = 54/82 (65%), Gaps = 5/82 (6%)
Query 1 SQCSVVATYWQTETGGHVLAPFP---GATVTKPGCAMVPFFGAEPAIVDPQSGEEKTGNN 57
S+C++V T+WQTETGG +AP P GA + P AM PFFG P ++D + G G+N
Sbjct 176 SRCTLVDTWWQTETGGICIAPRPSEEGAEIL-PAMAMRPFFGIVPVLMD-EKGSVVEGSN 233
Query 58 VCGVLCIKRSWPGMCRTVHRAH 79
V G LCI ++WPGM RT++ H
Sbjct 234 VSGALCISQAWPGMARTIYGDH 255
> Hs8923896
Length=701
Score = 75.9 bits (185), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 38/79 (48%), Positives = 48/79 (60%), Gaps = 2/79 (2%)
Query 1 SQCSVVATYWQTETGGHVLAPFPGATVTKPGCAMVPFFGAEPAIVDPQSGEEKTGNNVCG 60
+C +V T+WQTETGGH+L P PGAT KPG A PFFG PAI++ E +
Sbjct 457 QRCPIVDTFWQTETGGHMLTPLPGATPMKPGSATFPFFGVAPAILNESGEELEGEAEG-- 514
Query 61 VLCIKRSWPGMCRTVHRAH 79
L K+ WPG+ RTV+ H
Sbjct 515 YLVFKQPWPGIMRTVYGNH 533
> Hs21269872
Length=606
Score = 75.9 bits (185), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 38/79 (48%), Positives = 48/79 (60%), Gaps = 2/79 (2%)
Query 1 SQCSVVATYWQTETGGHVLAPFPGATVTKPGCAMVPFFGAEPAIVDPQSGEEKTGNNVCG 60
+C +V T+WQTETGGH+L P PGAT KPG A PFFG PAI++ E +
Sbjct 362 QRCPIVDTFWQTETGGHMLTPLPGATPMKPGSATFPFFGVAPAILNESGEELEGEAEG-- 419
Query 61 VLCIKRSWPGMCRTVHRAH 79
L K+ WPG+ RTV+ H
Sbjct 420 YLVFKQPWPGIMRTVYGNH 438
> ECU05g0310
Length=632
Score = 70.9 bits (172), Expect = 6e-13, Method: Composition-based stats.
Identities = 33/76 (43%), Positives = 44/76 (57%), Gaps = 3/76 (3%)
Query 3 CSVVATYWQTETGGHVLAPFPGATVTKPGCAMVPFFGAEPAIVDPQSGEE---KTGNNVC 59
C V+ TYWQTE+GG ++AP P A KP CA +PF G E I DP S + +
Sbjct 387 CPVIDTYWQTESGGILIAPIPHAVDIKPECACLPFLGQEVVIADPSSRSDCVVEAKAYEL 446
Query 60 GVLCIKRSWPGMCRTV 75
G + +K SWPG+ R +
Sbjct 447 GRVLLKGSWPGITRGI 462
> CE20020
Length=67
Score = 30.4 bits (67), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 20/69 (28%), Positives = 28/69 (40%), Gaps = 9/69 (13%)
Query 1 SQCSVVATY-WQTETGGHVLAPFPGATVTKPGCAMVPFFGAEPAIVDPQSGEEKTGNNVC 59
S VA Y W+T+ G + C ++ G P + D QS T
Sbjct 5 STAKTVAPYTWKTDAGARA--------TSTSDCIVLVLNGNNPIVADVQSCNADTTLPAL 56
Query 60 GVLCIKRSW 68
GV+C KR+W
Sbjct 57 GVMCGKRAW 65
> Hs22044164
Length=476
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 19/61 (31%), Positives = 27/61 (44%), Gaps = 1/61 (1%)
Query 18 VLAPFPGATVTKPGCAMVPFFGAEPAIVDPQSGEEKTGN-NVCGVLCIKRSWPGMCRTVH 76
V++ PG + PG A P A PA P+SG + + + C V C + VH
Sbjct 146 VISDSPGVRIWVPGAAPAPNSRAHPAGAAPESGSPRCPDASPCPVSCRPQDPRSTLAKVH 205
Query 77 R 77
R
Sbjct 206 R 206
> YDR264c
Length=764
Score = 27.3 bits (59), Expect = 6.4, Method: Composition-based stats.
Identities = 18/52 (34%), Positives = 26/52 (50%), Gaps = 6/52 (11%)
Query 13 ETGGHVLAPFPGATVTKPG-----CAMVPFFGAEPAIVDPQSG-EEKTGNNV 58
E+ VLAP PG+T+ KP C V A++ G ++ TG+NV
Sbjct 643 ESNDTVLAPVPGSTIRKPRTCFGVCYAVTGMDQWLAVIKETIGIKDSTGHNV 694
Lambda K H
0.320 0.135 0.462
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1168763848
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40