bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2178_orf1
Length=67
Score E
Sequences producing significant alignments: (Bits) Value
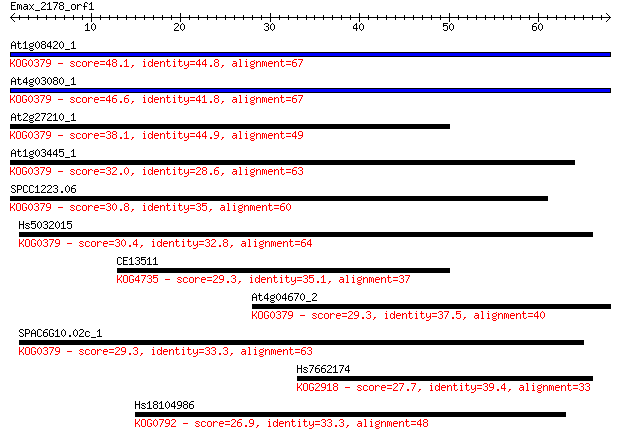
At1g08420_1 48.1 4e-06
At4g03080_1 46.6 1e-05
At2g27210_1 38.1 0.004
At1g03445_1 32.0 0.28
SPCC1223.06 30.8 0.63
Hs5032015 30.4 0.88
CE13511 29.3 1.8
At4g04670_2 29.3 1.8
SPAC6G10.02c_1 29.3 2.0
Hs7662174 27.7 5.4
Hs18104986 26.9 8.0
> At1g08420_1
Length=665
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 30/68 (44%), Positives = 40/68 (58%), Gaps = 2/68 (2%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILH-G 59
VV G G LS E+L++L L ++ +W V +QG PG RYGHVM ++ G
Sbjct 200 VVIQGGIGPAGLSAEDLHVLDLTQQ-RPRWHRVVVQGPGPGPRYGHVMALVGQRYLMAIG 258
Query 60 GNDGERPL 67
GNDG+RPL
Sbjct 259 GNDGKRPL 266
> At4g03080_1
Length=533
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 28/68 (41%), Positives = 38/68 (55%), Gaps = 2/68 (2%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMG-YSTPNIILHG 59
VVF G G S ++LY+L + + +W V +QG PG RYGHVM S ++
Sbjct 111 VVFQGGIGPAGHSTDDLYVLDMTND-KFKWHRVVVQGDGPGPRYGHVMDLVSQRYLVTVT 169
Query 60 GNDGERPL 67
GNDG+R L
Sbjct 170 GNDGKRAL 177
> At2g27210_1
Length=384
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 22/49 (44%), Positives = 29/49 (59%), Gaps = 1/49 (2%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMG 49
VV G G LS E+L++L L ++ +W V +QG PG RYGHVM
Sbjct 165 VVIQGGIGPAGLSAEDLHVLDLTQQ-RPRWHRVVVQGPGPGPRYGHVMA 212
> At1g03445_1
Length=472
Score = 32.0 bits (71), Expect = 0.28, Method: Compositional matrix adjust.
Identities = 18/64 (28%), Positives = 32/64 (50%), Gaps = 4/64 (6%)
Query 1 VVFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPN-IILHG 59
++ G G S ++Y+L + +W+ + G TP RYGHVM + +++
Sbjct 161 ILIQGGIGPSGPSDGDVYMLDMTNN---KWIKFLVGGETPSPRYGHVMDIAAQRWLVIFS 217
Query 60 GNDG 63
GN+G
Sbjct 218 GNNG 221
> SPCC1223.06
Length=1147
Score = 30.8 bits (68), Expect = 0.63, Method: Composition-based stats.
Identities = 21/62 (33%), Positives = 26/62 (41%), Gaps = 5/62 (8%)
Query 1 VVFGGATGGGSLSPEE--LYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILH 58
+VFGG T ++ LYLL L W G+ P RYGH + I L
Sbjct 147 IVFGGLTNHDVADRQDNSLYLLNTSS---LVWQKANASGARPSGRYGHTISCLGSKICLF 203
Query 59 GG 60
GG
Sbjct 204 GG 205
> Hs5032015
Length=372
Score = 30.4 bits (67), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 29/64 (45%), Gaps = 2/64 (3%)
Query 2 VFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILHGGN 61
VFGG G P + L + L W G+ P R+GHVM + + +HGG
Sbjct 154 VFGGGERGAQ--PVQDTKLHVFDANTLTWSQPETLGNPPSPRHGHVMVAAGTKLFIHGGL 211
Query 62 DGER 65
G+R
Sbjct 212 AGDR 215
> CE13511
Length=506
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 13/37 (35%), Positives = 21/37 (56%), Gaps = 1/37 (2%)
Query 13 SPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMG 49
SP +LY+ L E + ++ VP++ PG Y H +G
Sbjct 107 SPHQLYI-NLSTEGNNEYTVVPVKDVRPGPTYSHTLG 142
> At4g04670_2
Length=506
Score = 29.3 bits (64), Expect = 1.8, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 18/40 (45%), Gaps = 0/40 (0%)
Query 28 LQWLAVPLQGSTPGRRYGHVMGYSTPNIILHGGNDGERPL 67
LQW V QG P R+ H M + GG +GE L
Sbjct 211 LQWKEVEQQGQWPCARHSHAMVAYGSQSFMFGGYNGENVL 250
> SPAC6G10.02c_1
Length=343
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 27/63 (42%), Gaps = 5/63 (7%)
Query 2 VFGGATGGGSLSPEELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILHGGN 61
V GG G LS +L+L L L W V G PG R GH + ++GG
Sbjct 262 VHGGHNDTGPLS--DLWLFDLET---LSWTEVRSIGRFPGPREGHQATTIDDTVYIYGGR 316
Query 62 DGE 64
D +
Sbjct 317 DNK 319
> Hs7662174
Length=686
Score = 27.7 bits (60), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 20/34 (58%), Gaps = 1/34 (2%)
Query 33 VPLQGSTPG-RRYGHVMGYSTPNIILHGGNDGER 65
V +G P +RYGH + +P++IL G GE+
Sbjct 346 VSSEGQVPNLKRYGHASVFLSPDVILSAGGFGEQ 379
> Hs18104986
Length=913
Score = 26.9 bits (58), Expect = 8.0, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 22/48 (45%), Gaps = 4/48 (8%)
Query 15 EELYLLGLRKEPHLQWLAVPLQGSTPGRRYGHVMGYSTPNIILHGGND 62
E+LY RK+P L L + RY V+ Y T ++L G D
Sbjct 651 EQLY----RKKPGLAITFAKLPQNLDKNRYKDVLPYDTTRVLLQGNED 694
Lambda K H
0.317 0.144 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1160559522
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40