bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2180_orf1
Length=104
Score E
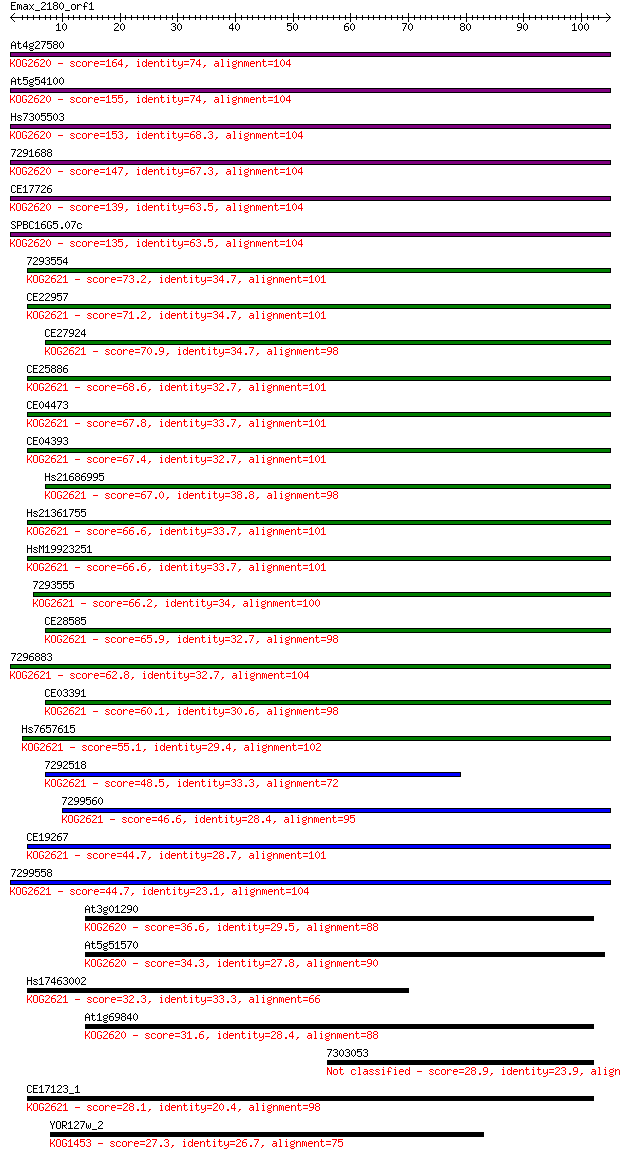
Sequences producing significant alignments: (Bits) Value
At4g27580 164 4e-41
At5g54100 155 2e-38
Hs7305503 153 7e-38
7291688 147 6e-36
CE17726 139 1e-33
SPBC16G5.07c 135 2e-32
7293554 73.2 1e-13
CE22957 71.2 4e-13
CE27924 70.9 6e-13
CE25886 68.6 3e-12
CE04473 67.8 5e-12
CE04393 67.4 6e-12
Hs21686995 67.0 8e-12
Hs21361755 66.6 1e-11
HsM19923251 66.6 1e-11
7293555 66.2 2e-11
CE28585 65.9 2e-11
7296883 62.8 1e-10
CE03391 60.1 1e-09
Hs7657615 55.1 3e-08
7292518 48.5 3e-06
7299560 46.6 1e-05
CE19267 44.7 4e-05
7299558 44.7 4e-05
At3g01290 36.6 0.011
At5g51570 34.3 0.054
Hs17463002 32.3 0.21
At1g69840 31.6 0.42
7303053 28.9 2.7
CE17123_1 28.1 3.8
YOR127w_2 27.3 7.8
> At4g27580
Length=515
Score = 164 bits (415), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 77/104 (74%), Positives = 88/104 (84%), Gaps = 0/104 (0%)
Query 1 KEEALTIPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELG 60
KEEA+ IPNQTAIT+DNV++ IDGVLYV+I D ASYGVE+PI+AV QLAQTTMRSELG
Sbjct 105 KEEAIPIPNQTAITKDNVSIHIDGVLYVKIVDPKLASYGVESPIYAVVQLAQTTMRSELG 164
Query 61 KLTLDNTFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
K+TLD TF ER+ LN+ IVEAIN AA WGL CLRYEIRDI+ P
Sbjct 165 KITLDKTFEERDTLNEKIVEAINVAAKDWGLQCLRYEIRDIMPP 208
> At5g54100
Length=401
Score = 155 bits (392), Expect = 2e-38, Method: Composition-based stats.
Identities = 77/104 (74%), Positives = 87/104 (83%), Gaps = 0/104 (0%)
Query 1 KEEALTIPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELG 60
KEEA+ I NQTAIT+DNV++ IDGVLYV+I D ASYGVENPI+AV QLAQTTMRSELG
Sbjct 148 KEEAIPIGNQTAITKDNVSIHIDGVLYVKIVDPKLASYGVENPIYAVMQLAQTTMRSELG 207
Query 61 KLTLDNTFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
K+TLD TF ER+ LN+ IVEAIN AA WGL CLRYEIRDI+ P
Sbjct 208 KITLDKTFEERDTLNEKIVEAINVAAKDWGLQCLRYEIRDIMPP 251
> Hs7305503
Length=356
Score = 153 bits (387), Expect = 7e-38, Method: Compositional matrix adjust.
Identities = 71/104 (68%), Positives = 86/104 (82%), Gaps = 0/104 (0%)
Query 1 KEEALTIPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELG 60
KE + +P Q+A+T DNVTLQIDGVLY++I D YKASYGVE+P +AVTQLAQTTMRSELG
Sbjct 80 KEIVINVPEQSAVTLDNVTLQIDGVLYLRIMDPYKASYGVEDPEYAVTQLAQTTMRSELG 139
Query 61 KLTLDNTFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
KL+LD F ER +LN +IV+AIN AA WG+ CLRYEI+DI +P
Sbjct 140 KLSLDKVFRERESLNASIVDAINQAADCWGIRCLRYEIKDIHVP 183
> 7291688
Length=323
Score = 147 bits (370), Expect = 6e-36, Method: Composition-based stats.
Identities = 70/104 (67%), Positives = 86/104 (82%), Gaps = 0/104 (0%)
Query 1 KEEALTIPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELG 60
KE A+ +P Q+AIT DNVTL IDGVLY++I D YKASYGVE+P FA+TQLAQTTMRSELG
Sbjct 42 KEIAIDVPKQSAITSDNVTLSIDGVLYLRIIDPYKASYGVEDPEFAITQLAQTTMRSELG 101
Query 61 KLTLDNTFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
K+++D F ER +LN +IV++IN A+ WG+ CLRYEIRDI LP
Sbjct 102 KMSMDKVFRERESLNVSIVDSINKASEAWGIACLRYEIRDIRLP 145
> CE17726
Length=334
Score = 139 bits (350), Expect = 1e-33, Method: Compositional matrix adjust.
Identities = 66/107 (61%), Positives = 83/107 (77%), Gaps = 3/107 (2%)
Query 1 KEEALTIPNQTAITRDNVTLQIDGVLYVQIKDAYKA---SYGVENPIFAVTQLAQTTMRS 57
+E A+ IP Q AIT DNV L++DGVLY+++ D YKA SYGV++P FAVTQLAQTTMRS
Sbjct 87 REIAIEIPEQGAITIDNVQLRLDGVLYLRVFDPYKACDASYGVDDPEFAVTQLAQTTMRS 146
Query 58 ELGKLTLDNTFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
E+GK+ LD F ER LN+ IV AIN A+ PWG+ C+RYEIRD+ +P
Sbjct 147 EVGKINLDTVFKERELLNENIVFAINKASAPWGIQCMRYEIRDMQMP 193
> SPBC16G5.07c
Length=354
Score = 135 bits (339), Expect = 2e-32, Method: Composition-based stats.
Identities = 66/104 (63%), Positives = 82/104 (78%), Gaps = 0/104 (0%)
Query 1 KEEALTIPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELG 60
KE AL IP Q+AIT DNV+L +DGVLY+Q+ D YKASYGVE+ +A++QLAQTTMRSE+G
Sbjct 95 KERALEIPTQSAITLDNVSLGLDGVLYIQVYDPYKASYGVEDADYAISQLAQTTMRSEIG 154
Query 61 KLTLDNTFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
+LTLD+ ER +LN I +AIN AA WG+ CLR+EIRDI P
Sbjct 155 RLTLDHVLRERQSLNIHITDAINKAAESWGIRCLRHEIRDIRPP 198
> 7293554
Length=339
Score = 73.2 bits (178), Expect = 1e-13, Method: Composition-based stats.
Identities = 35/101 (34%), Positives = 58/101 (57%), Gaps = 0/101 (0%)
Query 4 ALTIPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELGKLT 63
+ +P Q +++D+VT+ +D V+Y +I D KA V N + + LA TT+R+ LG
Sbjct 74 SFDVPPQEVLSKDSVTVTVDAVVYYRISDPLKAVIQVYNYSHSTSLLAATTLRNVLGTRN 133
Query 64 LDNTFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
L ER ++ T+ +++ A PWG+ R EI+D+ LP
Sbjct 134 LSELLTERETISHTMQMSLDEATDPWGVKVERVEIKDVSLP 174
> CE22957
Length=281
Score = 71.2 bits (173), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 35/101 (34%), Positives = 60/101 (59%), Gaps = 0/101 (0%)
Query 4 ALTIPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELGKLT 63
+ +P Q +++D+VT+ +D V+Y +I +A + VE+ + LAQTT+R+ LG T
Sbjct 96 SFDVPPQEILSKDSVTVSVDAVIYFRISNATVSVINVEDAARSTKLLAQTTLRNFLGTRT 155
Query 64 LDNTFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
L R+A++ + A++ A PWG+ R EI+D+ LP
Sbjct 156 LAEMLSSRDAISMQMQAALDEATDPWGVKVERVEIKDVRLP 196
> CE27924
Length=330
Score = 70.9 bits (172), Expect = 6e-13, Method: Composition-based stats.
Identities = 34/98 (34%), Positives = 58/98 (59%), Gaps = 0/98 (0%)
Query 7 IPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELGKLTLDN 66
+P+Q ++RD+VT+ +D V+Y ++ D + GV N + LAQTT+R+ LG TL
Sbjct 112 VPSQEILSRDSVTVSVDAVVYFKVFDPITSVVGVGNATDSTKLLAQTTLRTILGTHTLSE 171
Query 67 TFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
+R ++ + +++ A PWG+ R E+RD+ LP
Sbjct 172 ILSDREKISADMKISLDEATEPWGIKVERVELRDVRLP 209
> CE25886
Length=367
Score = 68.6 bits (166), Expect = 3e-12, Method: Composition-based stats.
Identities = 33/101 (32%), Positives = 57/101 (56%), Gaps = 0/101 (0%)
Query 4 ALTIPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELGKLT 63
+ +P Q ++RD+VT+ ++ V+Y ++ + + V + F+ LAQTT+R+ LG T
Sbjct 180 SFDVPPQEILSRDSVTVSVEAVIYFRVSNPVISVTNVNDAQFSTRLLAQTTLRNVLGTKT 239
Query 64 LDNTFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
L ER+A+ + ++ PWG+ R EI+DI LP
Sbjct 240 LSEMLSERDAIASISEKVLDEGTDPWGVKVERVEIKDIRLP 280
> CE04473
Length=280
Score = 67.8 bits (164), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 34/101 (33%), Positives = 57/101 (56%), Gaps = 0/101 (0%)
Query 4 ALTIPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELGKLT 63
+ ++P Q +T+D+VT +D V+Y +I +A + VEN + LAQTT+R+ LG +
Sbjct 105 SFSVPPQEILTKDSVTTSVDAVIYYRISNATVSVANVENAHHSTRLLAQTTLRNMLGTRS 164
Query 64 LDNTFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
L +R L ++ ++ A WG+ R EI+D+ LP
Sbjct 165 LSEILSDRETLAASMQTILDEATESWGIKVERVEIKDVRLP 205
> CE04393
Length=481
Score = 67.4 bits (163), Expect = 6e-12, Method: Composition-based stats.
Identities = 33/101 (32%), Positives = 59/101 (58%), Gaps = 0/101 (0%)
Query 4 ALTIPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELGKLT 63
+ +P Q +++D+VT+ +D V+Y +I +A + VE+ + LAQTT+R+ LG T
Sbjct 187 SFEVPPQEILSKDSVTVAVDAVVYFRISNATISVTNVEDAARSTKLLAQTTLRNILGTKT 246
Query 64 LDNTFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
L +R A++ + ++ A PWG+ R E++D+ LP
Sbjct 247 LAEMLSDREAISHQMQTTLDEATEPWGVKVERVEVKDVRLP 287
> Hs21686995
Length=291
Score = 67.0 bits (162), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 38/98 (38%), Positives = 54/98 (55%), Gaps = 0/98 (0%)
Query 7 IPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELGKLTLDN 66
IP Q +TRD+VT Q+DGV+Y +I A A V + A LAQTT+R+ LG TL
Sbjct 100 IPPQEILTRDSVTTQVDGVVYYRIYSAVSAVANVNDVHQATFLLAQTTLRNVLGTQTLSQ 159
Query 67 TFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
R + +I ++ A WG+ R EI+D+ +P
Sbjct 160 ILAGREEIAHSIQTLLDDATELWGIRVARVEIKDVRIP 197
> Hs21361755
Length=288
Score = 66.6 bits (161), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 34/101 (33%), Positives = 56/101 (55%), Gaps = 0/101 (0%)
Query 4 ALTIPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELGKLT 63
+ IP Q +T+D+VT+ +DGV+Y ++++A A + N A LAQTT+R+ LG
Sbjct 100 SFDIPPQEILTKDSVTISVDGVVYYRVQNATLAVANITNADSATRLLAQTTLRNVLGTKN 159
Query 64 LDNTFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
L +R + + ++ A WG+ R EI+D+ LP
Sbjct 160 LSQILSDREEIAHNMQSTLDDATDAWGIKVERVEIKDVKLP 200
> HsM19923251
Length=288
Score = 66.6 bits (161), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 34/101 (33%), Positives = 56/101 (55%), Gaps = 0/101 (0%)
Query 4 ALTIPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELGKLT 63
+ IP Q +T+D+VT+ +DGV+Y ++++A A + N A LAQTT+R+ LG
Sbjct 100 SFDIPPQEILTKDSVTISVDGVVYYRVQNATLAVANITNADSATRLLAQTTLRNVLGTKN 159
Query 64 LDNTFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
L +R + + ++ A WG+ R EI+D+ LP
Sbjct 160 LSQILSDREEIAHNMQSTLDDATDAWGIKVERVEIKDVKLP 200
> 7293555
Length=350
Score = 66.2 bits (160), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 34/100 (34%), Positives = 53/100 (53%), Gaps = 0/100 (0%)
Query 5 LTIPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELGKLTL 64
+P Q +T+D+VT+ +D V+Y +I D A VE+ + LA TT+R+ +G L
Sbjct 135 FNVPQQEMLTKDSVTVTVDAVVYYRISDPLYAVIQVEDYSMSTRLLAATTLRNIVGTRNL 194
Query 65 DNTFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
ER L + ++ A PWG+ R EI+D+ LP
Sbjct 195 SELLTERETLAHNMQATLDEATEPWGVMVERVEIKDVSLP 234
> CE28585
Length=285
Score = 65.9 bits (159), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 32/98 (32%), Positives = 55/98 (56%), Gaps = 0/98 (0%)
Query 7 IPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELGKLTLDN 66
+P Q +++D+VT+ +D V+Y + D + V++ I++ LAQTT+R+ LG TL
Sbjct 104 VPPQEILSKDSVTVSVDAVVYFRTSDPIASVNNVDDAIYSTKLLAQTTLRNALGMKTLTE 163
Query 67 TFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
ER A+ + ++ WG+ R E++DI LP
Sbjct 164 MLTEREAIAQLCETILDEGTEHWGVKVERVEVKDIRLP 201
> 7296883
Length=225
Score = 62.8 bits (151), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 34/104 (32%), Positives = 54/104 (51%), Gaps = 0/104 (0%)
Query 1 KEEALTIPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELG 60
+ + + Q +TRD VT+ IDGV+Y IK + A V +P A +LA TT+R+ G
Sbjct 41 RTRSFDLHRQEILTRDMVTISIDGVVYYSIKSPFDAMLQVYDPEEATEKLAMTTLRNVAG 100
Query 61 KLTLDNTFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
L + + L+ I + + PWG+ R EI++I +P
Sbjct 101 THKLMDLLSSKEYLSNQIEGILYNSTEPWGIRVERVEIKEIFMP 144
> CE03391
Length=267
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 30/98 (30%), Positives = 54/98 (55%), Gaps = 0/98 (0%)
Query 7 IPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELGKLTLDN 66
+P Q +TRD+VT+ +D +Y + D + V + + QLAQ+++R+ LG +L
Sbjct 88 VPTQEMLTRDSVTIGVDAAVYYRTSDPIASLARVNDAHMSTRQLAQSSLRNVLGTRSLAE 147
Query 67 TFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
+R+ + + +++A WG+ R EI+DI LP
Sbjct 148 LMTDRHGIAVQVKYILDSATLFWGIHVERVEIKDIRLP 185
> Hs7657615
Length=383
Score = 55.1 bits (131), Expect = 3e-08, Method: Composition-based stats.
Identities = 30/102 (29%), Positives = 53/102 (51%), Gaps = 0/102 (0%)
Query 3 EALTIPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELGKL 62
+ L IP +T+D ++ID + Y ++++A + + AV L QTTM+ L
Sbjct 170 QTLEIPFHEIVTKDMFIMEIDAICYYRMENASLLLSSLAHVSKAVQFLVQTTMKRLLAHR 229
Query 63 TLDNTFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
+L LER ++ + A+++ WG+ R EI+D+ LP
Sbjct 230 SLTEILLERKSIAQDAKVALDSVTCIWGIKVERIEIKDVRLP 271
> 7292518
Length=382
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 42/72 (58%), Gaps = 0/72 (0%)
Query 7 IPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELGKLTLDN 66
+P Q +T+D+VT+ +D V+Y ++ +A + VEN + LAQTT+R+ +G L
Sbjct 60 VPPQEVLTKDSVTVSVDAVVYYRVSNATVSIANVENAHHSTRLLAQTTLRNTMGTRHLHE 119
Query 67 TFLERNALNKTI 78
ER ++ T+
Sbjct 120 ILSERMTISGTM 131
> 7299560
Length=368
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/95 (28%), Positives = 50/95 (52%), Gaps = 0/95 (0%)
Query 10 QTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELGKLTLDNTFL 69
Q +T+D+VT+ ++ V+Y I + V++ A ++Q T+R+ +G TL+
Sbjct 43 QDVLTKDSVTITVNAVVYYCIYSPIDSIIQVDDAKQATQLISQVTLRNIVGSKTLNVLLT 102
Query 70 ERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
R L++ I +A+ WG+ R ++ DI LP
Sbjct 103 SRQQLSREIQQAVAGITYRWGVRVERVDVMDITLP 137
> CE19267
Length=298
Score = 44.7 bits (104), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 29/101 (28%), Positives = 57/101 (56%), Gaps = 0/101 (0%)
Query 4 ALTIPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELGKLT 63
+ +P Q +++D VT+ +D V++ +I +A + +E+ + LAQTT+R+ LG T
Sbjct 101 SFEVPPQELLSKDAVTVAVDAVVFFRISNATISVINIEDAARSTKLLAQTTLRNILGTKT 160
Query 64 LDNTFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
L +R+ ++ + ++ PWG+ R E++D+ LP
Sbjct 161 LTEMLSDRDVISLQMQATLDETTIPWGVKVERVEMKDVRLP 201
> 7299558
Length=474
Score = 44.7 bits (104), Expect = 4e-05, Method: Composition-based stats.
Identities = 24/104 (23%), Positives = 53/104 (50%), Gaps = 0/104 (0%)
Query 1 KEEALTIPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELG 60
+ + + + Q +T+D+V++ ++ V++ I D + V++ A +++Q T+R+ +G
Sbjct 93 RTDVVNVDPQEMLTKDSVSITVNAVVFYCIYDPINSIIKVDDARDATERISQVTLRNIVG 152
Query 61 KLTLDNTFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDILLP 104
L R L+ I +A+ WG+ R ++ +I LP
Sbjct 153 SKGLHELLASRQQLSLEIQQAVAKITERWGVRVERVDLMEISLP 196
> At3g01290
Length=285
Score = 36.6 bits (83), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 26/90 (28%), Positives = 46/90 (51%), Gaps = 2/90 (2%)
Query 14 TRDNVTLQI-DGVLYVQIKD-AYKASYGVENPIFAVTQLAQTTMRSELGKLTLDNTFLER 71
T+DNV + + + Y + D A A Y + NP + +R+ + KL LD+ F ++
Sbjct 62 TKDNVFVTVVASIQYRVLADKASDAFYRLSNPTTQIKAYVFDVIRACVPKLNLDDVFEQK 121
Query 72 NALNKTIVEAINTAAGPWGLTCLRYEIRDI 101
N + K++ E ++ A +G L+ I DI
Sbjct 122 NEIAKSVEEELDKAMTAYGYEILQTLIIDI 151
> At5g51570
Length=292
Score = 34.3 bits (77), Expect = 0.054, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 46/92 (50%), Gaps = 7/92 (7%)
Query 14 TRDNVTLQ-IDGVLYVQIK-DAYKASYGVENPIFAVTQLAQTTMRSELGKLTLDNTFLER 71
T+DNV +Q + + Y +K A A Y ++NP + +R+ + +TLD F ++
Sbjct 64 TKDNVFVQLVCSIQYRVVKASADDAFYELQNPKEQIQAYVFDVVRALVPMMTLDALFEQK 123
Query 72 NALNKTIVEAINTAAGPWGLTCLRYEIRDILL 103
+ K+++E + G +G Y I IL+
Sbjct 124 GEVAKSVLEELEKVMGAYG-----YSIEHILM 150
> Hs17463002
Length=115
Score = 32.3 bits (72), Expect = 0.21, Method: Compositional matrix adjust.
Identities = 22/73 (30%), Positives = 40/73 (54%), Gaps = 8/73 (10%)
Query 4 ALTIPNQTAITRDNVTLQIDGVLYVQIKDAYKA--SYGVENPIFAVTQLAQTTMRSEL-- 59
+L IP + +T+D+VT+++D + + QI++A A + PIF +L+Q +E+
Sbjct 14 SLDIPPEEKLTKDSVTIRVDNLCH-QIQNATMADITNADSKPIFRHQELSQVHSDTEIAY 72
Query 60 ---GKLTLDNTFL 69
G L D +F
Sbjct 73 NMSGSLDNDMSFF 85
> At1g69840
Length=286
Score = 31.6 bits (70), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 44/92 (47%), Gaps = 6/92 (6%)
Query 14 TRDNVTLQIDGVLYVQIK----DAYKASYGVENPIFAVTQLAQTTMRSELGKLTLDNTFL 69
T+DNV + + V +Q + A A Y + N + +R+ + KL LD+TF
Sbjct 62 TKDNVFVTV--VASIQYRALAESAQDAFYKLSNTRNQIQAYVFDVIRASVPKLDLDSTFE 119
Query 70 ERNALNKTIVEAINTAAGPWGLTCLRYEIRDI 101
++N + KT+ + A +G ++ I DI
Sbjct 120 QKNDIAKTVETELEKAMSHYGYEIVQTLIVDI 151
> 7303053
Length=430
Score = 28.9 bits (63), Expect = 2.7, Method: Composition-based stats.
Identities = 11/46 (23%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 56 RSELGKLTLDNTFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDI 101
R+ +G +T++ + +R +K + E ++ G+T + Y I+D+
Sbjct 112 RAIMGSMTVEEIYKDRKKFSKQVFEVASSDLANMGITVVSYTIKDL 157
> CE17123_1
Length=324
Score = 28.1 bits (61), Expect = 3.8, Method: Composition-based stats.
Identities = 20/100 (20%), Positives = 47/100 (47%), Gaps = 2/100 (2%)
Query 4 ALTIPNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELGKLT 63
A +P IT D +++ ++++I+D A GV++ +V LA T + + K
Sbjct 132 AFNVPPLQIITTDRGLVELGATVFLKIRDPIAAVCGVQDRNASVRTLANTMLYRYISKKR 191
Query 64 LDN--TFLERNALNKTIVEAINTAAGPWGLTCLRYEIRDI 101
+ + + +R ++ + + + + +G+ EI D+
Sbjct 192 ICDVTSSQDRRIISANLKDELGSFTCQFGVEITDVEISDV 231
> YOR127w_2
Length=884
Score = 27.3 bits (59), Expect = 7.8, Method: Composition-based stats.
Identities = 20/75 (26%), Positives = 34/75 (45%), Gaps = 5/75 (6%)
Query 8 PNQTAITRDNVTLQIDGVLYVQIKDAYKASYGVENPIFAVTQLAQTTMRSELGKLTLDNT 67
PN ++ N + + VL+ + K G IF +++Q M L + L+N
Sbjct 191 PNNSS---RNASELLTSVLHSPVSVNMKNPKGSNTDIFNTGEISQ--MDPSLSRKVLNNI 245
Query 68 FLERNALNKTIVEAI 82
E NAL + +VE +
Sbjct 246 VEETNALQRPVVEVV 260
Lambda K H
0.317 0.134 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1174332180
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40