bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2196_orf1
Length=143
Score E
Sequences producing significant alignments: (Bits) Value
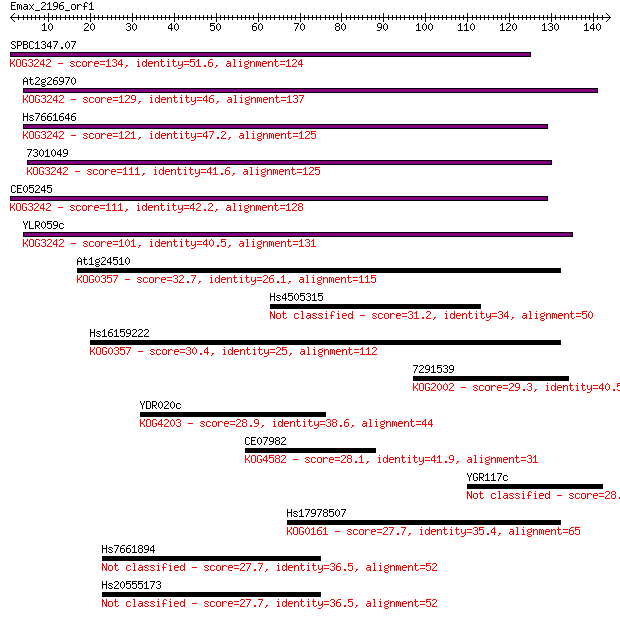
SPBC1347.07 134 8e-32
At2g26970 129 2e-30
Hs7661646 121 6e-28
7301049 111 5e-25
CE05245 111 5e-25
YLR059c 101 4e-22
At1g24510 32.7 0.25
Hs4505315 31.2 0.76
Hs16159222 30.4 1.2
7291539 29.3 2.8
YDR020c 28.9 3.0
CE07982 28.1 5.3
YGR117c 28.1 5.8
Hs17978507 27.7 7.5
Hs7661894 27.7 8.4
Hs20555173 27.7 8.4
> SPBC1347.07
Length=180
Score = 134 bits (336), Expect = 8e-32, Method: Compositional matrix adjust.
Identities = 64/124 (51%), Positives = 87/124 (70%), Gaps = 0/124 (0%)
Query 1 KKELEEMGSWCREHHGKSGLTQACLRSCMTASDAEQRIIAFLKLNGVGAKEAVLAGNSVH 60
+K+L EM WC E HGKSGLT+ C +S +T D E +++A++K +EA++AGNSVH
Sbjct 52 EKQLSEMNDWCIEQHGKSGLTERCRQSNLTVKDVENQLLAYIKKYIPKKREALIAGNSVH 111
Query 61 MDKEFLRREMPELIEFLHYRILDISSIKVVAKSWFPRVAPPKKRYRHRALADIQESIEEL 120
D FL EMP++IE LHYRI+D+S+IK +AK W P + K+ HRAL+DI ESI EL
Sbjct 112 ADVRFLSVEMPKIIEHLHYRIIDVSTIKELAKRWCPDIPAYDKKGDHRALSDILESIGEL 171
Query 121 AYYR 124
+YR
Sbjct 172 QHYR 175
> At2g26970
Length=227
Score = 129 bits (324), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 63/139 (45%), Positives = 93/139 (66%), Gaps = 3/139 (2%)
Query 4 LEEMGSWCREHHGKSGLTQACLRSCMTASDAEQRIIAFLKLNGVGAKEAVLAGNSVHMDK 63
L++M WC+ HHG SGLT+ L S +T +AEQ++I F+K + VG+ +LAGNSV++D
Sbjct 74 LDKMDDWCQTHHGASGLTKKVLLSAITEREAEQKVIEFVKKH-VGSGNPLLAGNSVYVDF 132
Query 64 EFLRREMPELIEFLHYRILDISSIKVVAKSWFP--RVAPPKKRYRHRALADIQESIEELA 121
FL++ MPEL + ++D+SS+K + WFP R P K+ HRA+ DI+ESI+EL
Sbjct 133 LFLKKYMPELAALFPHILVDVSSVKALCARWFPIERRKAPAKKNNHRAMDDIRESIKELK 192
Query 122 YYRKQIFRDLEDSLTSRDN 140
YY+K IF+ L + R+N
Sbjct 193 YYKKTIFKKLGTKVQKREN 211
> Hs7661646
Length=237
Score = 121 bits (303), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 59/127 (46%), Positives = 82/127 (64%), Gaps = 3/127 (2%)
Query 4 LEEMGSWCREHHGKSGLTQACLRSCMTASDAEQRIIAFLKLNGVGAKEAVLAGNSVHMDK 63
L+ M WC+EHHG+SGLT+A S +T AE ++F++ LAGNSVH DK
Sbjct 90 LDSMSDWCKEHHGRSGLTKAVKESTITLQQAEYEFLSFVRQQ-TPPGLCPLAGNSVHEDK 148
Query 64 EFLRREMPELIEFLHYRILDISSIKVVAKSWFPRVA--PPKKRYRHRALADIQESIEELA 121
+FL + MP+ ++ LHYRI+D+S++K + + W+P PKK HRAL DI ESI+EL
Sbjct 149 KFLDKYMPQFMKHLHYRIIDVSTVKELCRRWYPEEYEFAPKKAASHRALDDISESIKELQ 208
Query 122 YYRKQIF 128
+YR IF
Sbjct 209 FYRNNIF 215
> 7301049
Length=211
Score = 111 bits (277), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 52/127 (40%), Positives = 82/127 (64%), Gaps = 3/127 (2%)
Query 5 EEMGSWCREHHGKSGLTQACLRSCMTASDAEQRIIAFLKLNGVGAKEAVLAGNSVHMDKE 64
+ M WC +HH SGL C S + +A ++++L+ N + + L GNSV+ D+
Sbjct 86 DSMNEWCMKHHYNSGLIDRCKSSDVNLEEASNLVLSYLEKN-IPKRACPLGGNSVYTDRL 144
Query 65 FLRREMPELIEFLHYRILDISSIKVVAKSWFPRV--APPKKRYRHRALADIQESIEELAY 122
F+ + MP + +LHYRI+D+S+IK +AK W P + + PKK + HR+L DI+ESI+ELAY
Sbjct 145 FIMKFMPLVDAYLHYRIVDVSTIKELAKRWHPAILDSAPKKSFTHRSLDDIRESIKELAY 204
Query 123 YRKQIFR 129
Y+ +F+
Sbjct 205 YKANLFK 211
> CE05245
Length=193
Score = 111 bits (277), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 54/128 (42%), Positives = 82/128 (64%), Gaps = 1/128 (0%)
Query 1 KKELEEMGSWCREHHGKSGLTQACLRSCMTASDAEQRIIAFLKLNGVGAKEAVLAGNSVH 60
K+ L+ M W R ++GL + + S + +DAE +I FLKL+ + K + AGNS++
Sbjct 59 KEVLDNMEEWPRNTFHENGLMEKIIASKYSMADAENEVIDFLKLHALPGKSPI-AGNSIY 117
Query 61 MDKEFLRREMPELIEFLHYRILDISSIKVVAKSWFPRVAPPKKRYRHRALADIQESIEEL 120
MD+ F+++ MP+L +F HYR +D+S+IK + + W+P PKK+ HRA DI ESI EL
Sbjct 118 MDRLFIKKYMPKLDKFAHYRCIDVSTIKGLVQRWYPDYKHPKKQCTHRAFDDIMESIAEL 177
Query 121 AYYRKQIF 128
YR+ IF
Sbjct 178 KNYRESIF 185
> YLR059c
Length=269
Score = 101 bits (252), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 53/133 (39%), Positives = 78/133 (58%), Gaps = 2/133 (1%)
Query 4 LEEMGSWCREHHGKSGLTQACLRSCMTASDAEQRIIAFLKLNGVGAKEAVLAGNSVHMDK 63
+ +M WC EHHG SGLT L S T + E ++ +++ VLAGNSVHMD+
Sbjct 109 MNKMNEWCIEHHGNSGLTAKVLASEKTLAQVEDELLEYIQRYIPDKNVGVLAGNSVHMDR 168
Query 64 EFLRREMPELIEFLHYRILDISSIKVVAKSWFPRVAP--PKKRYRHRALADIQESIEELA 121
F+ RE P++I+ L YRI+D+SSI VA+ P + PKK H A +DI+ESI +L
Sbjct 169 LFMVREFPKVIDHLFYRIVDVSSIMEVARRHNPALQARNPKKEAAHTAYSDIKESIAQLQ 228
Query 122 YYRKQIFRDLEDS 134
+Y + +++
Sbjct 229 WYMDNYLKPPQET 241
> At1g24510
Length=535
Score = 32.7 bits (73), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 30/119 (25%), Positives = 51/119 (42%), Gaps = 16/119 (13%)
Query 17 KSGLTQACLRSCMTASDAEQRIIAF--LKLNG-VGAK-EAVLAGNSVHMDKEFLRREMPE 72
K L + +++ + +D E+R + +K+ G VG K E + +DK+ +MP+
Sbjct 178 KRSLAEIAVKAVLAVADLERRDVNLDLIKVEGKVGGKLEDTELIYGILIDKDMSHPQMPK 237
Query 73 LIEFLHYRILDISSIKVVAKSWFPRVAPPKKRYRHRALADIQESIEELAYYRKQIFRDL 131
IE H IL PPK + +H+ D E E L +Q F ++
Sbjct 238 QIEDAHIAILTCP------------FEPPKPKTKHKVDIDTVEKFETLRKQEQQYFDEM 284
> Hs4505315
Length=1465
Score = 31.2 bits (69), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 17/50 (34%), Positives = 27/50 (54%), Gaps = 5/50 (10%)
Query 63 KEFLRREMPELIEFLHYRILDISSIKVVAKSWFPRVAPPKKRYRHRALAD 112
KEFLR++ P E+LH+ + + +++V K VA KK + L D
Sbjct 1122 KEFLRKQGPHFAEYLHWDVTEECEVRLVCK-----VANTKKETVFKWLKD 1166
> Hs16159222
Length=541
Score = 30.4 bits (67), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 28/116 (24%), Positives = 51/116 (43%), Gaps = 16/116 (13%)
Query 20 LTQACLRSCMTASDAEQRIIAF--LKLNG-VGAK-EAVLAGNSVHMDKEFLRREMPELIE 75
+ + + + +T +D E+R + F +K+ G VG + E V +DK+F +MP+ +E
Sbjct 185 MAEIAVNAVLTVADMERRDVDFELIKVEGKVGGRLEDTKLIKGVIVDKDFSHPQMPKKVE 244
Query 76 FLHYRILDISSIKVVAKSWFPRVAPPKKRYRHRALADIQESIEELAYYRKQIFRDL 131
IL PPK + +H+ E + L Y K+ F ++
Sbjct 245 DAKIAILTCP------------FEPPKPKTKHKLDVTSVEDYKALQKYEKEKFEEM 288
> 7291539
Length=894
Score = 29.3 bits (64), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 20/37 (54%), Gaps = 0/37 (0%)
Query 97 RVAPPKKRYRHRALADIQESIEELAYYRKQIFRDLED 133
+ PPKK+ R RA QE IE L + + F D +D
Sbjct 854 KSGPPKKKTRKRAAGKEQEDIEPLKKPKSKEFIDTDD 890
> YDR020c
Length=232
Score = 28.9 bits (63), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 27/44 (61%), Gaps = 2/44 (4%)
Query 32 SDAEQRIIAFLKLNGVGAKEAVLAGNSVHMDKEFLRREMPELIE 75
SDA++R+I+ +K VG+ E + + +MD LR EM + IE
Sbjct 122 SDADKRLISLIKKKNVGSNEQLAQLITEYMDH--LRPEMQQYIE 163
> CE07982
Length=498
Score = 28.1 bits (61), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 20/31 (64%), Gaps = 2/31 (6%)
Query 57 NSVHMDKEFLRREMPELIEFLHYRILDISSI 87
N++ + F ++P+L FLH RILD+S I
Sbjct 84 NNIRPELSFYHSQLPQL--FLHVRILDVSVI 112
> YGR117c
Length=476
Score = 28.1 bits (61), Expect = 5.8, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 21/38 (55%), Gaps = 6/38 (15%)
Query 110 LADIQESIEELAYYRKQIFRD------LEDSLTSRDNA 141
L D+Q +E+ YY+++ F+D + D L DNA
Sbjct 57 LEDLQSVVEDRIYYKRRSFKDRFKTLSINDDLAPIDNA 94
> Hs17978507
Length=1581
Score = 27.7 bits (60), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 23/69 (33%), Positives = 32/69 (46%), Gaps = 5/69 (7%)
Query 67 RREMPELIEFLHYRILDISSIKVVAKSWFPRVAPPKKRYRHRALADIQESIEELAYYRK- 125
R+ M IE LH +I DI+ K + R+ K ++R L + QE + EL K
Sbjct 1248 RKAMEVEIEDLHLQIDDIAKAKTALEEQLSRLQREKNEIQNR-LEEDQEDMNELMKKHKA 1306
Query 126 ---QIFRDL 131
Q RDL
Sbjct 1307 AVAQASRDL 1315
> Hs7661894
Length=1698
Score = 27.7 bits (60), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 31/64 (48%), Gaps = 12/64 (18%)
Query 23 ACLRSCMTASDAEQRIIAFLK------------LNGVGAKEAVLAGNSVHMDKEFLRREM 70
ACLRS T + Q +I FL LN +GA++A+ H+ K L +E+
Sbjct 713 ACLRSPNTDREVLQELIFFLHRLTSVSRDYAVVLNQLGARDAISKALEKHLGKLELAQEL 772
Query 71 PELI 74
+++
Sbjct 773 RDMV 776
> Hs20555173
Length=1698
Score = 27.7 bits (60), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 31/64 (48%), Gaps = 12/64 (18%)
Query 23 ACLRSCMTASDAEQRIIAFLK------------LNGVGAKEAVLAGNSVHMDKEFLRREM 70
ACLRS T + Q +I FL LN +GA++A+ H+ K L +E+
Sbjct 713 ACLRSPNTDREVLQELIFFLHRLTSVSRDYAVVLNQLGARDAISKALEKHLGKLELAQEL 772
Query 71 PELI 74
+++
Sbjct 773 RDMV 776
Lambda K H
0.320 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1639544670
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40