bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2217_orf1
Length=114
Score E
Sequences producing significant alignments: (Bits) Value
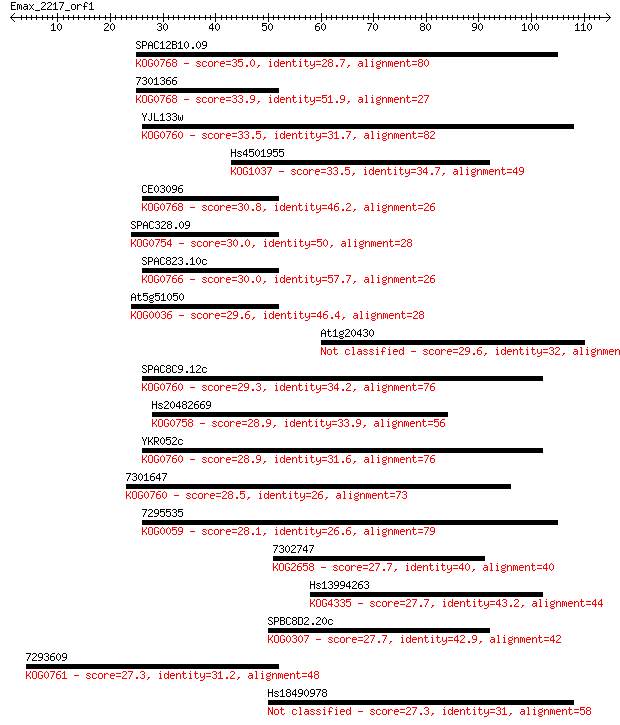
SPAC12B10.09 35.0 0.038
7301366 33.9 0.072
YJL133w 33.5 0.10
Hs4501955 33.5 0.11
CE03096 30.8 0.56
SPAC328.09 30.0 0.95
SPAC823.10c 30.0 1.2
At5g51050 29.6 1.3
At1g20430 29.6 1.4
SPAC8C9.12c 29.3 2.0
Hs20482669 28.9 2.1
YKR052c 28.9 2.5
7301647 28.5 3.2
7295535 28.1 4.4
7302747 27.7 5.4
Hs13994263 27.7 6.0
SPBC8D2.20c 27.7 6.1
7293609 27.3 7.5
Hs18490978 27.3 7.7
> SPAC12B10.09
Length=345
Score = 35.0 bits (79), Expect = 0.038, Method: Composition-based stats.
Identities = 23/80 (28%), Positives = 31/80 (38%), Gaps = 11/80 (13%)
Query 25 ALAAGASTGIIADISLFPLDFLKTHAQGNNVPPVSAAAAPAGPAPPASSQAPVAAAASVA 84
AL AG G+ D+SLFP+D LKT Q A G S+
Sbjct 85 ALGAGICAGLAVDLSLFPIDTLKTRLQ-----------AKGGFVKNGGFHGVYRGLGSIL 133
Query 85 TGSPPSSRVNFRSSLEFRGR 104
GS P + + F + + R
Sbjct 134 VGSAPGASLFFTTYENMKSR 153
> 7301366
Length=297
Score = 33.9 bits (76), Expect = 0.072, Method: Compositional matrix adjust.
Identities = 14/27 (51%), Positives = 19/27 (70%), Gaps = 0/27 (0%)
Query 25 ALAAGASTGIIADISLFPLDFLKTHAQ 51
AL AG G++ DI+LFP+D +KT Q
Sbjct 30 ALVAGGVAGMVVDIALFPIDTVKTRLQ 56
> YJL133w
Length=314
Score = 33.5 bits (75), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 26/85 (30%), Positives = 41/85 (48%), Gaps = 4/85 (4%)
Query 26 LAAGASTGIIADISLFPLDFLKTHAQGNNVPPVSAAAAPAGPAPPASSQAPVA---AAAS 82
L AGA GI+ +FP+D LKT Q N +SA + + ++S+ +A S
Sbjct 37 LIAGAFAGIMEHSVMFPIDALKTRIQSANAKSLSAKNMLSQISHISTSEGTLALWKGVQS 96
Query 83 VATGSPPSSRVNFRSSLEFRGRTYV 107
V G+ P+ V F + EF + +
Sbjct 97 VILGAGPAHAVYF-GTYEFCKKNLI 120
> Hs4501955
Length=1014
Score = 33.5 bits (75), Expect = 0.11, Method: Composition-based stats.
Identities = 17/49 (34%), Positives = 26/49 (53%), Gaps = 1/49 (2%)
Query 43 LDFLKTHAQGNNVPPVSAAAAPAGPAPPASSQAPVAAAASVATGSPPSS 91
L LK Q PP ++A+ A P PP+++ AP A +S + P S+
Sbjct 345 LKKLKVKKQDRIFPPETSASVAATP-PPSTASAPAAVNSSASADKPLSN 392
> CE03096
Length=269
Score = 30.8 bits (68), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 26 LAAGASTGIIADISLFPLDFLKTHAQ 51
L GA+ G+ DI L+PLD +K+ Q
Sbjct 12 LVCGATAGLAVDIGLYPLDTIKSRMQ 37
> SPAC328.09
Length=298
Score = 30.0 bits (66), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 14/28 (50%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 24 VALAAGASTGIIADISLFPLDFLKTHAQ 51
V AAGA GI ++L+PLD +KT Q
Sbjct 10 VTFAAGAVAGISEVLTLYPLDVVKTRMQ 37
> SPAC823.10c
Length=296
Score = 30.0 bits (66), Expect = 1.2, Method: Compositional matrix adjust.
Identities = 15/26 (57%), Positives = 17/26 (65%), Gaps = 0/26 (0%)
Query 26 LAAGASTGIIADISLFPLDFLKTHAQ 51
LAAGA G I+ +L PLD LKT Q
Sbjct 18 LAAGALGGFISSTTLQPLDLLKTRCQ 43
> At5g51050
Length=487
Score = 29.6 bits (65), Expect = 1.3, Method: Composition-based stats.
Identities = 13/28 (46%), Positives = 17/28 (60%), Gaps = 0/28 (0%)
Query 24 VALAAGASTGIIADISLFPLDFLKTHAQ 51
V L AG G +A S++PLD +KT Q
Sbjct 305 VRLFAGGMAGAVAQASIYPLDLVKTRLQ 332
> At1g20430
Length=116
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 16/50 (32%), Positives = 27/50 (54%), Gaps = 0/50 (0%)
Query 60 AAAAPAGPAPPASSQAPVAAAASVATGSPPSSRVNFRSSLEFRGRTYVLL 109
A++AP G P + + P A +VA+G + + N +S +F T +LL
Sbjct 2 ASSAPQGSVDPLTGKDPAKALTAVASGFFENVKKNKQSFFQFAAMTGILL 51
> SPAC8C9.12c
Length=303
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 26/80 (32%), Positives = 37/80 (46%), Gaps = 5/80 (6%)
Query 26 LAAGASTGIIADISLFPLDFLKTHAQGNNVPPVSAAAAPAGPAPPASSQAPVAA----AA 81
L AGA +GI+ ++P+D +KT Q N S + SS V + +
Sbjct 23 LLAGAFSGILEHSVMYPVDAIKTRMQMLNGVSRSVSGNIVNSVIKISSTEGVYSLWRGIS 82
Query 82 SVATGSPPSSRVNFRSSLEF 101
SV G+ PS + F S LEF
Sbjct 83 SVIMGAGPSHAIYF-SVLEF 101
> Hs20482669
Length=164
Score = 28.9 bits (63), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 19/56 (33%), Positives = 24/56 (42%), Gaps = 6/56 (10%)
Query 28 AGASTGIIADISLFPLDFLKTHAQGNNVPPVSAAAAPAGPAPPASSQAPVAAAASV 83
AG + G + L P D +K Q P A P +PP Q PV AAS+
Sbjct 63 AGCTGGFLQAYCLAPFDLIKVRLQNQTEP----RAQPG--SPPPRYQGPVHCAASI 112
> YKR052c
Length=304
Score = 28.9 bits (63), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 24/79 (30%), Positives = 33/79 (41%), Gaps = 4/79 (5%)
Query 26 LAAGASTGIIADISLFPLDFLKTHAQG---NNVPPVSAAAAPAGPAPPASSQAPVAAAAS 82
L AGA GI+ +FP+D LKT Q N + + + S A S
Sbjct 27 LLAGAFAGIMEHSLMFPIDALKTRVQAAGLNKAASTGMISQISKISTMEGSMALWKGVQS 86
Query 83 VATGSPPSSRVNFRSSLEF 101
V G+ P+ V F + EF
Sbjct 87 VILGAGPAHAVYF-GTYEF 104
> 7301647
Length=379
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 19/76 (25%), Positives = 36/76 (47%), Gaps = 4/76 (5%)
Query 23 AVALAAGASTGIIADISLFPLDFLKTHAQGNNVPPVSAAAAPAGPAPPASSQA---PVAA 79
V + AGA G++ + ++PLD +KT Q + PP + + + P+
Sbjct 15 GVNMTAGAIAGVLEHVVMYPLDSVKTRMQSLS-PPTKNMNIVSTLRTMITREGLLRPIRG 73
Query 80 AASVATGSPPSSRVNF 95
A++V G+ P+ + F
Sbjct 74 ASAVVLGAGPAHSLYF 89
> 7295535
Length=1500
Score = 28.1 bits (61), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 21/79 (26%), Positives = 36/79 (45%), Gaps = 5/79 (6%)
Query 26 LAAGASTGIIADISLFPLDFLKTHAQGNNVPPVSAAAAPAGPAPPASSQAPVAAAASVAT 85
L + + G I+D+ L L+ + ++ AGP PAS ++ V A ++ T
Sbjct 640 LQSQVTAGSISDVELTTSSSLEQEFE-----QLNRNGEDAGPRRPASDRSGVVAGPAMIT 694
Query 86 GSPPSSRVNFRSSLEFRGR 104
PP++ FR + R R
Sbjct 695 DEPPTACKQFRLLMGKRLR 713
> 7302747
Length=511
Score = 27.7 bits (60), Expect = 5.4, Method: Composition-based stats.
Identities = 16/44 (36%), Positives = 21/44 (47%), Gaps = 4/44 (9%)
Query 51 QGNNVPPVSAAAAP----AGPAPPASSQAPVAAAASVATGSPPS 90
QG + V A + +GPA PA S P +A GSPP+
Sbjct 29 QGRTIVTVDAKESTPPKGSGPATPAGSPPPKNVSAPPTGGSPPA 72
> Hs13994263
Length=464
Score = 27.7 bits (60), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 19/52 (36%), Positives = 27/52 (51%), Gaps = 8/52 (15%)
Query 58 VSAAAAPAGP--------APPASSQAPVAAAASVATGSPPSSRVNFRSSLEF 101
+S AA PAGP + P+SS APV GS SSR+ S++++
Sbjct 371 LSQAAEPAGPQDSATGSPSDPSSSLAPVQRPKLRRQGSVVSSRIQHLSTIDY 422
> SPBC8D2.20c
Length=1224
Score = 27.7 bits (60), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 18/44 (40%), Positives = 26/44 (59%), Gaps = 3/44 (6%)
Query 50 AQGNNVPPVSAAAAPAGPAPPASSQAPVAAAASVAT--GSPPSS 91
A G VPPV + P P P + Q+PVAAA+ +++ PPS+
Sbjct 940 ASGTRVPPVQQPSHP-NPYTPVAPQSPVAAASRISSSPNMPPSN 982
> 7293609
Length=443
Score = 27.3 bits (59), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 15/48 (31%), Positives = 21/48 (43%), Gaps = 6/48 (12%)
Query 4 SVSRASQVNETLMIFSRDEAVALAAGASTGIIADISLFPLDFLKTHAQ 51
++ RA V E +FS GA +G +A P D + TH Q
Sbjct 297 AIKRAFSVTEPTFLFS------FLTGAISGAVATFVTMPFDLITTHTQ 338
> Hs18490978
Length=559
Score = 27.3 bits (59), Expect = 7.7, Method: Composition-based stats.
Identities = 18/58 (31%), Positives = 29/58 (50%), Gaps = 2/58 (3%)
Query 50 AQGNNVPPVSAAAAPAGPAPPASSQAPVAAAASVATGSPPSSRVNFRSSLEFRGRTYV 107
A G ++PP A +PAGPA + A A++A + S +N +L R R ++
Sbjct 33 AGGGDLPP--APLSPAGPAAYSPPGPGPAPPAAMALRNDLGSNINVLKTLNLRFRCFL 88
Lambda K H
0.317 0.127 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1181971906
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40