bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2281_orf1
Length=161
Score E
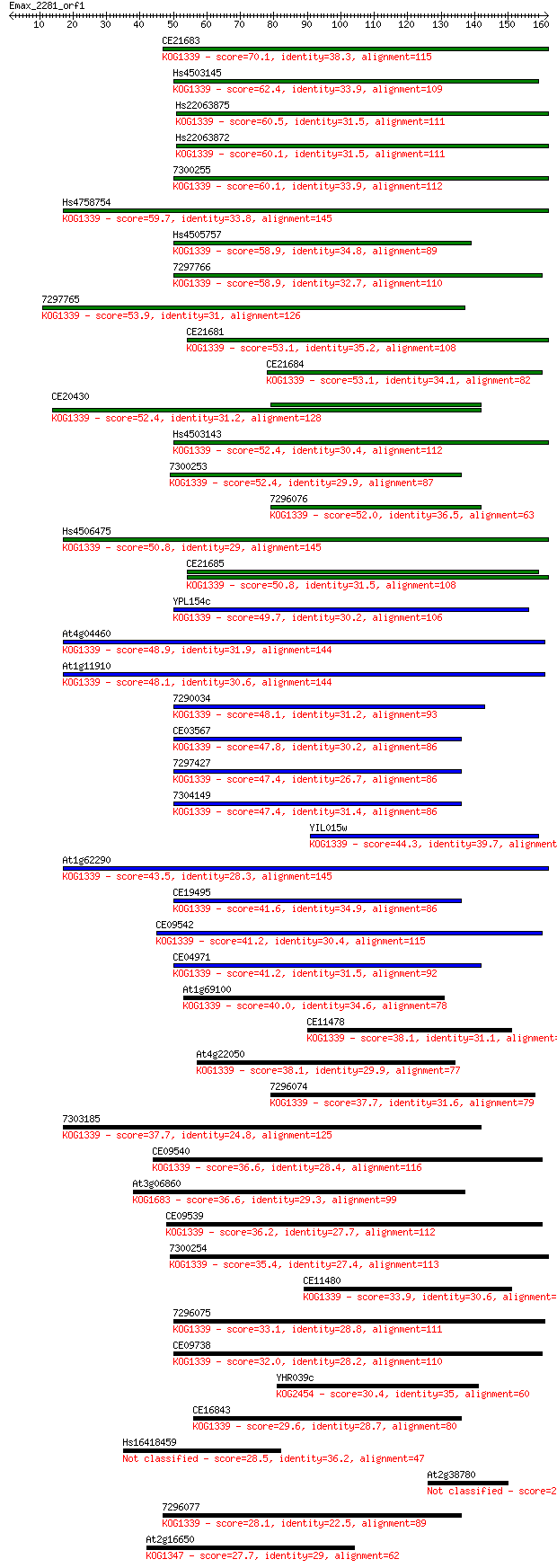
Sequences producing significant alignments: (Bits) Value
CE21683 70.1 2e-12
Hs4503145 62.4 4e-10
Hs22063875 60.5 2e-09
Hs22063872 60.1 2e-09
7300255 60.1 2e-09
Hs4758754 59.7 2e-09
Hs4505757 58.9 4e-09
7297766 58.9 4e-09
7297765 53.9 1e-07
CE21681 53.1 2e-07
CE21684 53.1 2e-07
CE20430 52.4 4e-07
Hs4503143 52.4 4e-07
7300253 52.4 4e-07
7296076 52.0 5e-07
Hs4506475 50.8 1e-06
CE21685 50.8 1e-06
YPL154c 49.7 3e-06
At4g04460 48.9 5e-06
At1g11910 48.1 6e-06
7290034 48.1 8e-06
CE03567 47.8 1e-05
7297427 47.4 1e-05
7304149 47.4 1e-05
YIL015w 44.3 1e-04
At1g62290 43.5 2e-04
CE19495 41.6 6e-04
CE09542 41.2 8e-04
CE04971 41.2 0.001
At1g69100 40.0 0.002
CE11478 38.1 0.007
At4g22050 38.1 0.007
7296074 37.7 0.009
7303185 37.7 0.011
CE09540 36.6 0.019
At3g06860 36.6 0.023
CE09539 36.2 0.030
7300254 35.4 0.048
CE11480 33.9 0.15
7296075 33.1 0.27
CE09738 32.0 0.48
YHR039c 30.4 1.5
CE16843 29.6 2.6
Hs16418459 28.5 5.3
At2g38780 28.5 5.4
7296077 28.1 7.5
At2g16650 27.7 9.9
> CE21683
Length=320
Score = 70.1 bits (170), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 44/118 (37%), Positives = 66/118 (55%), Gaps = 12/118 (10%)
Query 47 RGGCAPHEKFDPKYSSTF-SPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIKVPNQ 105
+ G A H KFD K SSTF S R + I YG+G+C + RD + +GG+K+ NQ
Sbjct 110 KSGFAKH-KFDTKASSTFVSETRKFS-------ISYGSGSCSGHLARDTITMGGLKIDNQ 161
Query 106 AIGLAVEESTHPFADLPFDGLVGLGFPDVSGEEGLP--SSALPIVDQTVKEKVLDRNV 161
G+A E FA+ P DG++GLG+P ++ ++ P + LP +D V +DR +
Sbjct 162 EFGVA-ETLASVFAEQPVDGILGLGWPALADDKVTPPMQNLLPQLDAKVFTVWMDRKL 218
> Hs4503145
Length=396
Score = 62.4 bits (150), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 37/110 (33%), Positives = 55/110 (50%), Gaps = 13/110 (11%)
Query 50 CAPHEKFDPKYSSTFSPIRSLTGDPAVAF-IQYGTGACVLRMGRDIVEIGGIKVPNQAIG 108
C H +F P SST+S P +F IQYGTG+ +G D V + G+ V Q G
Sbjct 114 CKTHSRFQPSQSSTYS-------QPGQSFSIQYGTGSLSGIIGADQVSVEGLTVVGQQFG 166
Query 109 LAVEESTHPFADLPFDGLVGLGFPDVSGEEGLPSSALPIVDQTVKEKVLD 158
+V E F D FDG++GLG+P ++ P+ D + + ++D
Sbjct 167 ESVTEPGQTFVDAEFDGILGLGYPSLA-----VGGVTPVFDNMMAQNLVD 211
> Hs22063875
Length=325
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 35/111 (31%), Positives = 55/111 (49%), Gaps = 11/111 (9%)
Query 51 APHEKFDPKYSSTFSPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGLA 110
A H +F+P+ SST+ I YGTG+ +G D V++GGI NQ GL+
Sbjct 50 ANHNRFNPEDSSTYQSTSETVS------ITYGTGSMTGILGYDTVQVGGISDTNQIFGLS 103
Query 111 VEESTHPFADLPFDGLVGLGFPDVSGEEGLPSSALPIVDQTVKEKVLDRNV 161
E PFDG++GL +P +S S A P+ D + ++ +++
Sbjct 104 ETEPGSFLYYAPFDGILGLAYPSISS-----SGATPVFDNIWNQGLVSQDL 149
> Hs22063872
Length=325
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 35/111 (31%), Positives = 55/111 (49%), Gaps = 11/111 (9%)
Query 51 APHEKFDPKYSSTFSPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGLA 110
A H +F+P+ SST+ I YGTG+ +G D V++GGI NQ GL+
Sbjct 50 ANHNRFNPEDSSTYQSTSETVS------ITYGTGSMTGILGYDTVQVGGISDTNQIFGLS 103
Query 111 VEESTHPFADLPFDGLVGLGFPDVSGEEGLPSSALPIVDQTVKEKVLDRNV 161
E PFDG++GL +P +S S A P+ D + ++ +++
Sbjct 104 ETEPGSFLYYAPFDGILGLAYPSISS-----SGATPVFDNIWNQGLVSQDL 149
> 7300255
Length=395
Score = 60.1 bits (144), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 38/112 (33%), Positives = 56/112 (50%), Gaps = 11/112 (9%)
Query 50 CAPHEKFDPKYSSTFSPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGL 109
C H +++ SSTF P D I YGTG+ R+ +D V IG + V NQ G+
Sbjct 119 CHHHHRYNASASSTFVP------DGRRFSIAYGTGSLSGRLAQDTVAIGQLVVQNQTFGM 172
Query 110 AVEESTHPFADLPFDGLVGLGFPDVSGEEGLPSSALPIVDQTVKEKVLDRNV 161
A E F D F G+VGLGF ++ E G+ P+ + ++++D V
Sbjct 173 ATHEPGPTFVDTNFAGIVGLGFRPIA-ELGIK----PLFESMCDQQLVDECV 219
> Hs4758754
Length=420
Score = 59.7 bits (143), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 49/145 (33%), Positives = 70/145 (48%), Gaps = 16/145 (11%)
Query 17 PQTFGSLQRNARSKNQFPIGLRSSFYLSTHRGGCAPHEKFDPKYSSTFSPIRSLTGDPAV 76
PQ F ++ + S N + R F+ C H +FDPK SS+F +
Sbjct 88 PQNF-TVAFDTGSSNLWVPSRRCHFFSVP----CWLHHRFDPKASSSFQ------ANGTK 136
Query 77 AFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGLAVEESTHPFADLPFDGLVGLGFPDVSG 136
IQYGTG + D + IGGIK + G A+ E + FA FDG++GLGFP +S
Sbjct 137 FAIQYGTGRVDGILSEDKLTIGGIKGASVIFGEALWEPSLVFAFAHFDGILGLGFPILSV 196
Query 137 EEGLPSSALPIVDQTVKEKVLDRNV 161
E P +D V++ +LD+ V
Sbjct 197 E-----GVRPPMDVLVEQGLLDKPV 216
> Hs4505757
Length=388
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 31/89 (34%), Positives = 47/89 (52%), Gaps = 6/89 (6%)
Query 50 CAPHEKFDPKYSSTFSPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGL 109
C H +F+P SST+S + +QYG+G+ G D + + I+VPNQ GL
Sbjct 109 CTSHSRFNPSESSTYST------NGQTFSLQYGSGSLTGFFGYDTLTVQSIQVPNQEFGL 162
Query 110 AVEESTHPFADLPFDGLVGLGFPDVSGEE 138
+ E F FDG++GL +P +S +E
Sbjct 163 SENEPGTNFVYAQFDGIMGLAYPALSVDE 191
> 7297766
Length=391
Score = 58.9 bits (141), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 36/110 (32%), Positives = 52/110 (47%), Gaps = 11/110 (10%)
Query 50 CAPHEKFDPKYSSTFSPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGL 109
C H K+D SST+ G+ I+YGTG+ + DIV I GI + NQ G
Sbjct 114 CQRHNKYDSSASSTYVA----NGEEFA--IEYGTGSLSGFLSNDIVTIAGISIQNQTFGE 167
Query 110 AVEESTHPFADLPFDGLVGLGFPDVSGEEGLPSSALPIVDQTVKEKVLDR 159
A+ E F D PF G++GL F ++ + P D + + +LD
Sbjct 168 ALSEPGTTFVDAPFAGILGLAFSAIAVD-----GVTPPFDNMISQGLLDE 212
> 7297765
Length=423
Score = 53.9 bits (128), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 39/126 (30%), Positives = 62/126 (49%), Gaps = 11/126 (8%)
Query 11 LCLIRVPQTFGSLQRNARSKNQFPIGLRSSFYLSTHRGGCAPHEKFDPKYSSTFSPIRSL 70
LC+ PQ F +LQ + S + + ++ S ST+ C H K++ SS S
Sbjct 78 LCIGTPPQCF-NLQFDTGSSDLWVPSVKCS---STNEA-CQKHNKYNSSASS------SH 126
Query 71 TGDPAVAFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGLAVEESTHPFADLPFDGLVGLG 130
D IQYG+G+ + D V+I G+ + NQ A++E F + FDG++G+
Sbjct 127 VEDGKGFSIQYGSGSLSGFLSTDTVDIDGMVIRNQTFAEAIDEPGSAFVNTIFDGIIGMA 186
Query 131 FPDVSG 136
F +SG
Sbjct 187 FASISG 192
> CE21681
Length=396
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 38/116 (32%), Positives = 61/116 (52%), Gaps = 19/116 (16%)
Query 54 EKFDPKYSSTF-SPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGLAVE 112
+KFD S+TF R + IQYG+G+C +G+D++ GG+ V +Q G+
Sbjct 115 QKFDTTKSTTFVKETRKFS-------IQYGSGSCNGYLGKDVLNFGGLTVQSQEFGV--- 164
Query 113 ESTH---PFADLPFDGLVGLGFPDVSGEEGLPSS----ALPIVDQTVKEKVLDRNV 161
STH F P DG++GLG+P ++ ++ +P A +D + LDRN+
Sbjct 165 -STHLADVFGYQPVDGILGLGWPALAVDQVVPPMQNLIAQKQLDAPLFTVWLDRNL 219
> CE21684
Length=435
Score = 53.1 bits (126), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 28/84 (33%), Positives = 46/84 (54%), Gaps = 3/84 (3%)
Query 78 FIQYGTGACVLRMGRDIVEIGGIKVPNQAIGLAVEESTHPFADLPFDGLVGLGFPDVSGE 137
FI Y TGAC +G D++++GG+ V NQ + + S P DG++GL +P V+ +
Sbjct 130 FINYPTGACSGYLGTDVLQLGGLTVKNQEFAVTTQISDQ-MGHFPLDGVLGLAWPPVAVD 188
Query 138 EGLP--SSALPIVDQTVKEKVLDR 159
+ P + +P +D + LDR
Sbjct 189 QITPPMQNLMPQLDAPIFTLWLDR 212
> CE20430
Length=638
Score = 52.4 bits (124), Expect = 4e-07, Method: Composition-based stats.
Identities = 26/63 (41%), Positives = 37/63 (58%), Gaps = 1/63 (1%)
Query 79 IQYGTGACVLRMGRDIVEIGGIKVPNQAIGLAVEESTHPFADLPFDGLVGLGFPDVSGEE 138
IQYG+G C +G D VEIGG+K+ Q G+A + F P DG+ GL +P +S ++
Sbjct 127 IQYGSGECSGYLGTDTVEIGGLKIQKQEFGVA-NIVDYDFGTRPIDGIFGLAWPALSVDQ 185
Query 139 GLP 141
P
Sbjct 186 VTP 188
Score = 32.3 bits (72), Expect = 0.36, Method: Composition-based stats.
Identities = 32/128 (25%), Positives = 57/128 (44%), Gaps = 10/128 (7%)
Query 14 IRVPQTFGSLQRNARSKNQFPIGLRSSFYLSTHRGGCAPHEKFDPKYSSTFSPIRSLTGD 73
I P S+ + S N + IG + + G H KF+ S++F + G+
Sbjct 460 IGTPPQSASVFMDTTSANWWVIGSKCTSANCNGYSGIRKH-KFNTTKSTSF-----VEGN 513
Query 74 PAVAFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGLAVEESTHPFADLPFDGLVGLGFPD 133
+ +YG C +G D V++GG+ + Q +G+A F P+ G+ L +P
Sbjct 514 RTFS-TEYGL--CTGYLGTDTVQMGGLTITKQELGIATIVGLG-FGLKPYVGIFELAWPA 569
Query 134 VSGEEGLP 141
+S ++ P
Sbjct 570 LSVDQVTP 577
> Hs4503143
Length=412
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 34/124 (27%), Positives = 64/124 (51%), Gaps = 24/124 (19%)
Query 50 CAPHEKFDPKYSSTFSPIRSLTGDPAVAF-IQYGTGACVLRMGRDIVEI----------- 97
C H K++ SST+ +++ T +F I YG+G+ + +D V +
Sbjct 117 CWIHHKYNSDKSSTY--VKNGT-----SFDIHYGSGSLSGYLSQDTVSVPCQSASSASAL 169
Query 98 GGIKVPNQAIGLAVEESTHPFADLPFDGLVGLGFPDVSGEEGLPSSALPIVDQTVKEKVL 157
GG+KV Q G A ++ F FDG++G+ +P +S ++ LP+ D +++K++
Sbjct 170 GGVKVERQVFGEATKQPGITFIAAKFDGILGMAYPRIS-----VNNVLPVFDNLMQQKLV 224
Query 158 DRNV 161
D+N+
Sbjct 225 DQNI 228
> 7300253
Length=465
Score = 52.4 bits (124), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 26/87 (29%), Positives = 44/87 (50%), Gaps = 6/87 (6%)
Query 49 GCAPHEKFDPKYSSTFSPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIKVPNQAIG 108
C H+++ P SST+ + + A I YG+G+ + +D V I G+ V NQ
Sbjct 185 ACKKHKQYHPAKSSTY-----VKNGKSFA-ITYGSGSVAGVLAKDTVRIAGLVVTNQTFA 238
Query 109 LAVEESTHPFADLPFDGLVGLGFPDVS 135
+ +E F FDG++GLG+ ++
Sbjct 239 MTTKEPGTTFVTSNFDGILGLGYRSIA 265
> 7296076
Length=405
Score = 52.0 bits (123), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 23/63 (36%), Positives = 37/63 (58%), Gaps = 0/63 (0%)
Query 79 IQYGTGACVLRMGRDIVEIGGIKVPNQAIGLAVEESTHPFADLPFDGLVGLGFPDVSGEE 138
IQYGTG+ + D V I G+ + NQ G AV + F D+ FDG++G+G+ ++ +
Sbjct 154 IQYGTGSVSGYLAIDTVTINGLAIANQTFGEAVSQPGASFTDVAFDGILGMGYQQIAEDN 213
Query 139 GLP 141
+P
Sbjct 214 VVP 216
> Hs4506475
Length=406
Score = 50.8 bits (120), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 42/145 (28%), Positives = 59/145 (40%), Gaps = 17/145 (11%)
Query 17 PQTFGSLQRNARSKNQFPIGLRSSFYLSTHRGGCAPHEKFDPKYSSTFSPIRSLTGDPAV 76
PQTF + S P S Y + C H+ FD SS++ +
Sbjct 96 PQTFKVVFDTGSSNVWVPSSKCSRLYTA-----CVYHKLFDASDSSSYKH------NGTE 144
Query 77 AFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGLAVEESTHPFADLPFDGLVGLGFPDVSG 136
++Y TG + +DI+ +GGI V Q G E PF FDG+VG+GF
Sbjct 145 LTLRYSTGTVSGFLSQDIITVGGITV-TQMFGEVTEMPALPFMLAEFDGVVGMGFI---- 199
Query 137 EEGLPSSALPIVDQTVKEKVLDRNV 161
E PI D + + VL +V
Sbjct 200 -EQAIGRVTPIFDNIISQGVLKEDV 223
> CE21685
Length=829
Score = 50.8 bits (120), Expect = 1e-06, Method: Composition-based stats.
Identities = 33/106 (31%), Positives = 53/106 (50%), Gaps = 14/106 (13%)
Query 54 EKFDPKYSSTF-SPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGLAVE 112
+KF+P SSTF R + IQYG+G +G D++++GG+ V Q G+A
Sbjct 115 QKFNPNKSSTFVKGTRRFS-------IQYGSGTSSGYLGTDVLQLGGLTVKAQEFGVATN 167
Query 113 ESTHPFADLPFDGLVGLGFPDVSGEEGLPSSALPIVDQTVKEKVLD 158
+ P DG+ GLG+P +S ++ P + + +K LD
Sbjct 168 LGS-VLGSEPMDGIFGLGWPAISVDQ-----VTPPMQNLISQKQLD 207
Score = 43.9 bits (102), Expect = 1e-04, Method: Composition-based stats.
Identities = 36/112 (32%), Positives = 56/112 (50%), Gaps = 12/112 (10%)
Query 54 EKFDPKYSSTFS-PIRSLTGDPAVAFIQYGTGACVLRMGRDIVEI-GGIKVPNQAIGLAV 111
+KFD SS+FS R + IQYG G C +G D V + GG+ + Q +G+A
Sbjct 550 QKFDTSKSSSFSRENRKFS-------IQYGKGLCSGYLGTDTVGLGGGLTIRKQELGIAN 602
Query 112 EESTHPFADLPFDGLVGLGFPDVSGEEGLP--SSALPIVDQTVKEKVLDRNV 161
+ FA P DG+ GL +P ++ ++ P + + +D V LDR +
Sbjct 603 KLDVD-FAVQPMDGIFGLAWPALAVDQITPPMQNLISQLDVPVFSVWLDRKI 653
> YPL154c
Length=405
Score = 49.7 bits (117), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 32/106 (30%), Positives = 51/106 (48%), Gaps = 6/106 (5%)
Query 50 CAPHEKFDPKYSSTFSPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGL 109
C H K+D + SS++ + IQYGTG+ + +D + IG + +P Q
Sbjct 127 CFLHSKYDHEASSSYK------ANGTEFAIQYGTGSLEGYISQDTLSIGDLTIPKQDFAE 180
Query 110 AVEESTHPFADLPFDGLVGLGFPDVSGEEGLPSSALPIVDQTVKEK 155
A E FA FDG++GLG+ +S ++ +P I + EK
Sbjct 181 ATSEPGLTFAFGKFDGILGLGYDTISVDKVVPPFYNAIQQDLLDEK 226
> At4g04460
Length=508
Score = 48.9 bits (115), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 46/148 (31%), Positives = 61/148 (41%), Gaps = 16/148 (10%)
Query 17 PQTFGSLQRNARSKNQFPIGLRSSFYLSTHRGGCAPHEKFDPKYSSTFSPIRSLTGDPAV 76
PQ F + S P + YLS C H K+ SS++ G PA
Sbjct 97 PQKFTVIFDTGSSNLWIP---STKCYLSV---ACYFHSKYKASQSSSYRK----NGKPAS 146
Query 77 AFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGLAVEESTHPFADLPFDGLVGLGFPDVSG 136
I+YGTGA D V++G I V Q A E F FDG++GLGF ++S
Sbjct 147 --IRYGTGAISGYFSNDDVKVGDIVVKEQEFIEATSEPGITFLLAKFDGILGLGFKEISV 204
Query 137 EEGLPSSALPIVDQTVKEKV----LDRN 160
P + VKE + L+RN
Sbjct 205 GNSTPVWYNMVEKGLVKEPIFSFWLNRN 232
> At1g11910
Length=506
Score = 48.1 bits (113), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 44/148 (29%), Positives = 60/148 (40%), Gaps = 16/148 (10%)
Query 17 PQTFGSLQRNARSKNQFPIGLRSSFYLSTHRGGCAPHEKFDPKYSSTFSPIRSLTGDPAV 76
PQ F + S P S Y S C H K+ SST+ +
Sbjct 92 PQKFTVVFDTGSSNLWVP---SSKCYFSL---ACLLHPKYKSSRSSTYEK------NGKA 139
Query 77 AFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGLAVEESTHPFADLPFDGLVGLGFPDVSG 136
A I YGTGA D V +G + V +Q A +E F FDG++GLGF ++S
Sbjct 140 AAIHYGTGAIAGFFSNDAVTVGDLVVKDQEFIEATKEPGITFVVAKFDGILGLGFQEISV 199
Query 137 EEGLPSSALPIVDQTVKEKV----LDRN 160
+ P + +KE V L+RN
Sbjct 200 GKAAPVWYNMLKQGLIKEPVFSFWLNRN 227
> 7290034
Length=407
Score = 48.1 bits (113), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 29/94 (30%), Positives = 43/94 (45%), Gaps = 8/94 (8%)
Query 50 CAPHEKFDPKYSSTFSPIRSLTGDPAVAF-IQYGTGACVLRMGRDIVEIGGIKVPNQAIG 108
C H+ F SST+ AF I YGTG+ + D V G+ + +Q G
Sbjct 113 CQDHQVFYKNKSSTYVA-------NGTAFSITYGTGSVSGYLSVDCVGFAGLTIQSQTFG 165
Query 109 LAVEESTHPFADLPFDGLVGLGFPDVSGEEGLPS 142
E F D FDG++G+GFP ++ + P+
Sbjct 166 EVTTEQGTNFVDAYFDGILGMGFPSLAVDGVTPT 199
> CE03567
Length=444
Score = 47.8 bits (112), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 26/86 (30%), Positives = 42/86 (48%), Gaps = 6/86 (6%)
Query 50 CAPHEKFDPKYSSTFSPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGL 109
C H ++D K SST+ D IQYGTG+ + +D V + G+ +Q
Sbjct 132 CMLHHRYDSKSSSTYKE------DGRKMAIQYGTGSMKGFISKDSVCVAGVCAEDQPFAE 185
Query 110 AVEESTHPFADLPFDGLVGLGFPDVS 135
A E F FDG++G+ +P+++
Sbjct 186 ATSEPGITFVAAKFDGILGMAYPEIA 211
> 7297427
Length=372
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 23/86 (26%), Positives = 41/86 (47%), Gaps = 6/86 (6%)
Query 50 CAPHEKFDPKYSSTFSPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGL 109
C H ++D SST+ + IQYGTG+ + D V++ G+ + +Q
Sbjct 105 CLTHNQYDSSASSTY------VANGESFSIQYGTGSLTGYLSTDTVDVNGLSIQSQTFAE 158
Query 110 AVEESTHPFADLPFDGLVGLGFPDVS 135
+ E F D FDG++G+ + ++
Sbjct 159 STNEPGTNFNDANFDGILGMAYESLA 184
> 7304149
Length=392
Score = 47.4 bits (111), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 27/86 (31%), Positives = 41/86 (47%), Gaps = 6/86 (6%)
Query 50 CAPHEKFDPKYSSTFSPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGL 109
C H K+D S T+ T + IQYG+G+ + D V I G+ + +Q
Sbjct 111 CLMHNKYDASKSKTY------TKNGTEFAIQYGSGSLSGYLSTDTVSIAGLDIKDQTFAE 164
Query 110 AVEESTHPFADLPFDGLVGLGFPDVS 135
A+ E F FDG++GLG+ +S
Sbjct 165 ALSEPGLVFVAAKFDGILGLGYNSIS 190
> YIL015w
Length=587
Score = 44.3 bits (103), Expect = 1e-04, Method: Composition-based stats.
Identities = 27/72 (37%), Positives = 38/72 (52%), Gaps = 10/72 (13%)
Query 91 GRDIVEIGGIKVPNQAIGLAVEESTHPFADLPFDGLVGLGFP---DVSGEEGLPSSALPI 147
G + V I GI +PN G+A +A P G++G+GFP V G EG P+ P
Sbjct 141 GTETVSINGIDIPNIQFGVA------KYATTPVSGVLGIGFPRRESVKGYEGAPNEYYPN 194
Query 148 VDQTVK-EKVLD 158
Q +K EK++D
Sbjct 195 FPQILKSEKIID 206
> At1g62290
Length=526
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 41/145 (28%), Positives = 61/145 (42%), Gaps = 17/145 (11%)
Query 17 PQTFGSLQRNARSKNQFPIGLRSSFYLSTHRGGCAPHEKFDPKYSSTFSPIRSLTGDPAV 76
PQ F + S P G + F LS C H K+ SST+ +G A
Sbjct 99 PQKFTVIFDTGSSNLWVPSG-KCFFSLS-----CYFHAKYKSSRSSTYKK----SGKRAA 148
Query 77 AFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGLAVEESTHPFADLPFDGLVGLGFPDVSG 136
I YG+G+ D V +G + V +Q E F FDGL+GLGF +++
Sbjct 149 --IHYGSGSISGFFSYDAVTVGDLVVKDQEFIETTSEPGLTFLVAKFDGLLGLGFQEIA- 205
Query 137 EEGLPSSALPIVDQTVKEKVLDRNV 161
+A P+ +K+ ++ R V
Sbjct 206 ----VGNATPVWYNMLKQGLIKRPV 226
> CE19495
Length=398
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 30/90 (33%), Positives = 39/90 (43%), Gaps = 10/90 (11%)
Query 50 CAPHEKFDPKYSSTFSPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIKV----PNQ 105
C H +FD K SS S T A IQYGTG+ + D+V G NQ
Sbjct 107 CRMHNRFDCKKSS------SCTATGASFEIQYGTGSMKGTVDNDVVCFGHDTTYCTDKNQ 160
Query 106 AIGLAVEESTHPFADLPFDGLVGLGFPDVS 135
+ A E F FDG+ G+G+ +S
Sbjct 161 GLACATSEPGITFVAAKFDGIFGMGWDTIS 190
> CE09542
Length=389
Score = 41.2 bits (95), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 35/121 (28%), Positives = 57/121 (47%), Gaps = 19/121 (15%)
Query 45 THRGGCAPHEKFDPKYSSTFSPI-RSLTGDPAVAFIQYGTGACVLRMGRDIVEIGG---- 99
T + C KFD SSTF +S T IQYG+G +G+D V G
Sbjct 101 TCKTNCKTKSKFDSTASSTFVKNGKSWT-------IQYGSGDAAGILGQDTVRFGAKGDS 153
Query 100 -IKVPNQAIGLAVEESTHPFADLPFDGLVGLGFPDVSGEEGLPSSALPIVDQTVKEKVLD 158
+ VP G+A + S F + DG++GL F ++ + +P P+++ + + +LD
Sbjct 154 QLSVPTTTFGIASKISAD-FKNDATDGILGLAFTSLAVDGVVP----PLIN-AINQGILD 207
Query 159 R 159
+
Sbjct 208 Q 208
> CE04971
Length=709
Score = 41.2 bits (95), Expect = 0.001, Method: Composition-based stats.
Identities = 29/97 (29%), Positives = 44/97 (45%), Gaps = 12/97 (12%)
Query 50 CAPHEKFDPKYSSTFSPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGI-----KVPN 104
C KFD SS++ + I+YGTG+ +G+D + GGI VPN
Sbjct 423 CFYQNKFDASKSSSYKT------NGRNFIIEYGTGSARGFLGQDTITFGGIGEPQLAVPN 476
Query 105 QAIGLAVEESTHPFADLPFDGLVGLGFPDVSGEEGLP 141
G A + F P DG++GL F ++ ++ P
Sbjct 477 TVFGQATSLAAF-FEGQPLDGILGLAFKSIAVDQITP 512
> At1g69100
Length=343
Score = 40.0 bits (92), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/78 (34%), Positives = 43/78 (55%), Gaps = 6/78 (7%)
Query 53 HEKFDPKYSSTFSPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGLAVE 112
H KFD S T ++ G+ +A Y TG+ V + +D V +GG+ + +Q + LA
Sbjct 86 HPKFDKDASKTCRLMKG--GEVNIA---YETGSVVGILAQDNVNVGGVVIKSQDLFLARN 140
Query 113 ESTHPFADLPFDGLVGLG 130
T+ F + FDG++GLG
Sbjct 141 PDTY-FRSVKFDGVIGLG 157
> CE11478
Length=383
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 19/63 (30%), Positives = 36/63 (57%), Gaps = 4/63 (6%)
Query 90 MGRDIVEIGGIKVPNQAIGLAVEESTHPFADLPFDGLVGLGFP--DVSGEEGLPSSALPI 147
+G+D ++ G K+ NQ G+ + + + + FDG++GLG+P ++G + LP
Sbjct 137 LGKDNIQFAGSKIQNQDFGIGTDAT--RLSGVTFDGVLGLGWPATALNGTSTTMQNLLPQ 194
Query 148 VDQ 150
+DQ
Sbjct 195 LDQ 197
> At4g22050
Length=336
Score = 38.1 bits (87), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 38/78 (48%), Gaps = 1/78 (1%)
Query 57 DPKYSSTFSPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGLAVEE-ST 115
+P+ S R+ + A ++YG G+ + D V +GGI + +Q V+
Sbjct 83 NPRNRYISSASRTFKENGTKAELKYGKGSLTGFLSVDTVTVGGISITSQTFIEGVKTPYK 142
Query 116 HPFADLPFDGLVGLGFPD 133
F +PFDG++GL F D
Sbjct 143 EFFKKMPFDGILGLRFTD 160
> 7296074
Length=393
Score = 37.7 bits (86), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 25/80 (31%), Positives = 39/80 (48%), Gaps = 6/80 (7%)
Query 79 IQYGTGACVLRMGRDIVEIGGIKVPNQAIGLA-VEESTHPFADLPFDGLVGLGFPDVSGE 137
I YG+G+ + D V G +K+ NQ IGL + +S FDG+ G F +S
Sbjct 146 ITYGSGSVKGIVASDNVGFGDLKIQNQGIGLVNISDSCS-----VFDGIAGFAFQQLSMT 200
Query 138 EGLPSSALPIVDQTVKEKVL 157
+ +PS I Q V++ +
Sbjct 201 KSVPSFQQMIDQQLVEQPIF 220
> 7303185
Length=404
Score = 37.7 bits (86), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 31/125 (24%), Positives = 52/125 (41%), Gaps = 11/125 (8%)
Query 17 PQTFGSLQRNARSKNQFPIGLRSSFYLSTHRGGCAPHEKFDPKYSSTFSPIRSLTGDPAV 76
PQTF + S P +S ++ C H ++ K S++ + GD
Sbjct 95 PQTFKVIFDTGSSNLWVPSATCASTMVA-----CRVHNRYFAKRSTS----HQVRGDHFA 145
Query 77 AFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGLAVEESTHPFADLPFDGLVGLGFPDVSG 136
I YG+G+ + D V + G+++ +Q A E F FDG+ GL + +S
Sbjct 146 --IHYGSGSLSGFLSTDTVRVAGLEIRDQTFAEATEMPGPIFLAAKFDGIFGLAYRSISM 203
Query 137 EEGLP 141
+ P
Sbjct 204 QRIKP 208
> CE09540
Length=395
Score = 36.6 bits (83), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 33/121 (27%), Positives = 57/121 (47%), Gaps = 17/121 (14%)
Query 44 STHRGGCAPHEKFDPKYSSTFSPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGG---- 99
S G C +FD S+TF + IQY +G+ MG D V GG
Sbjct 102 SNCDGTCKGKSEFDSSKSTTFKK------NGQSWQIQYESGSAKGIMGVDTVAFGGASEQ 155
Query 100 -IKVPNQAIGLAVEESTHPFADLPFDGLVGLGFPDVSGEEGLPSSALPIVDQTVKEKVLD 158
+ VP+ G+A S++ F + +G++GL F ++ + P P+++ + +K+LD
Sbjct 156 QLLVPSTTFGIASHISSN-FKNDAAEGILGLAFTSLAVDGVTP----PLIN-AINKKILD 209
Query 159 R 159
+
Sbjct 210 Q 210
> At3g06860
Length=725
Score = 36.6 bits (83), Expect = 0.023, Method: Compositional matrix adjust.
Identities = 29/102 (28%), Positives = 43/102 (42%), Gaps = 17/102 (16%)
Query 38 RSSFYLSTHRGGCAPHEKFDPKYSSTFSPIRSLTG---DPAVAFIQYGTGACVLRMGRDI 94
R FYL + P DP+ RS++G DP +A + +DI
Sbjct 582 RKGFYLYDDKRKAKP----DPELKKYIEKARSISGVKLDPKLANLSE----------KDI 627
Query 95 VEIGGIKVPNQAIGLAVEESTHPFADLPFDGLVGLGFPDVSG 136
+E+ V N+A + E ADL G++G+GFP G
Sbjct 628 IEMTFFPVVNEACRVFAEGIAVKAADLDIAGIMGMGFPPYRG 669
> CE09539
Length=393
Score = 36.2 bits (82), Expect = 0.030, Method: Compositional matrix adjust.
Identities = 31/117 (26%), Positives = 54/117 (46%), Gaps = 17/117 (14%)
Query 48 GGCAPHEKFDPKYSSTFSPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIK-----V 102
G C ++ SSTF G P IQYG+G +G D V G + V
Sbjct 106 GSCKGKREYQSTKSSTFKA----NGKPWQ--IQYGSGNAKGYLGEDTVAFGAVTEKQLPV 159
Query 103 PNQAIGLAVEESTHPFADLPFDGLVGLGFPDVSGEEGLPSSALPIVDQTVKEKVLDR 159
P+ G+A S+ F + +G++GL F ++ + +P P+++ + + +LD+
Sbjct 160 PSTTFGIATHISSD-FKNDAAEGILGLAFTSLAVDHVVP----PLIN-AINQGLLDQ 210
> 7300254
Length=309
Score = 35.4 bits (80), Expect = 0.048, Method: Compositional matrix adjust.
Identities = 31/114 (27%), Positives = 58/114 (50%), Gaps = 13/114 (11%)
Query 49 GCAPHEKFDPKYSSTFSPI-RSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIKVPNQAI 107
C H K++ SS++ P R+ T ++YG+G V + +D + I G ++P+
Sbjct 40 ACQNHRKYNSSRSSSYIPDGRNFT-------LRYGSGMVVGYLSKDTMHIAGAELPHFTF 92
Query 108 GLAVEESTHPFADLPFDGLVGLGFPDVSGEEGLPSSALPIVDQTVKEKVLDRNV 161
G ++ F+ + FDGLVGLG +S S+ P ++ +++L++ V
Sbjct 93 GESLFLQHFAFSSVKFDGLVGLGLGVLSW-----SNTTPFLELLCAQRLLEKCV 141
> CE11480
Length=474
Score = 33.9 bits (76), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 19/64 (29%), Positives = 34/64 (53%), Gaps = 4/64 (6%)
Query 89 RMGRDIVEIGGIKVPNQAIGLAVEESTHPFADLPFDGLVGLGFP--DVSGEEGLPSSALP 146
+G+D + G + +Q G+ +T F + FDG++GLG+P ++G + LP
Sbjct 227 NVGKDTAQFAGFTIQSQDFGIGTA-ATRLFGE-TFDGVLGLGWPATALNGTSTTMQNLLP 284
Query 147 IVDQ 150
+DQ
Sbjct 285 QLDQ 288
> 7296075
Length=410
Score = 33.1 bits (74), Expect = 0.27, Method: Compositional matrix adjust.
Identities = 32/122 (26%), Positives = 49/122 (40%), Gaps = 18/122 (14%)
Query 50 CAPHEKFDPKYSSTFSPIRSLTGDPAVAF-IQYGT----GACVLR--MGRDIVEIGGIKV 102
CA K++ S+T+ I + AF I YG+ G L +D V G +
Sbjct 128 CANRIKYNSSASTTYRAINT-------AFNIAYGSNSEEGPIALSGFQSQDTVNFAGYSI 180
Query 103 PNQAIGLAVEESTHPFADLPFDGLVGLGFPDVSGEEGLP----SSALPIVDQTVKEKVLD 158
NQ F DG++GLGF ++ P A +V+++V L+
Sbjct 181 KNQIFAEITNAPETAFLKSQLDGILGLGFASIAINSITPPFYNLMAQGLVNRSVFSIYLN 240
Query 159 RN 160
RN
Sbjct 241 RN 242
> CE09738
Length=389
Score = 32.0 bits (71), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 31/115 (26%), Positives = 53/115 (46%), Gaps = 17/115 (14%)
Query 50 CAPHEKFDPKYSSTFSPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGG-----IKVPN 104
C F+ K SST+ T P I+Y +G +G D V+IGG + +P
Sbjct 107 CEKKRLFNEKASSTYIA----TNRPWQ--IKYASGDAYGTLGIDTVKIGGSGEAQLAIPR 160
Query 105 QAIGLAVEESTHPFADLPFDGLVGLGFPDVSGEEGLPSSALPIVDQTVKEKVLDR 159
+G+A + F P +G+ GL F ++ + P PI++ + + +LD+
Sbjct 161 SYLGVA-DTVGSDFKWSPKEGIFGLAFTALAVDNITP----PIIN-AINQGLLDQ 209
> YHR039c
Length=644
Score = 30.4 bits (67), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 29/61 (47%), Gaps = 3/61 (4%)
Query 81 YGTGACVLRMGRDIVEIGGIKVPNQAIGLAVEE-STHPFADLPFDGLVGLGFPDVSGEEG 139
+G G V G DI E + Q +A+ + +T LPF G+ G G+ GEEG
Sbjct 520 FGLGGSVF--GADIKECNYVANSLQTGNVAINDFATFYVCQLPFGGINGSGYGKFGGEEG 577
Query 140 L 140
L
Sbjct 578 L 578
> CE16843
Length=394
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 36/81 (44%), Gaps = 3/81 (3%)
Query 56 FDPKYSSTFSPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIKVPNQAIGLAVEEST 115
+ KY T S G P I YGTG+ G+D + + ++ +
Sbjct 113 YKKKYFGTDSSSYEKDGTPFS--ISYGTGSASGYFGKDTLCFSDTTLCIKSQIFGQANNI 170
Query 116 HPF-ADLPFDGLVGLGFPDVS 135
PF A+ DG++GLGF D++
Sbjct 171 APFFANQEIDGILGLGFTDLA 191
> Hs16418459
Length=263
Score = 28.5 bits (62), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 17/57 (29%), Positives = 27/57 (47%), Gaps = 10/57 (17%)
Query 35 IGLRSSFYLSTHRGGCAPHEKFDPKYSS----------TFSPIRSLTGDPAVAFIQY 81
+G SF+L+ G + HE P+ + P R+LTG+ +V F+QY
Sbjct 8 LGFLISFFLTGVAGTQSTHESLKPQRVQFQSRNFHNILQWQPGRALTGNSSVYFVQY 64
> At2g38780
Length=435
Score = 28.5 bits (62), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 11/24 (45%), Positives = 15/24 (62%), Gaps = 0/24 (0%)
Query 126 LVGLGFPDVSGEEGLPSSALPIVD 149
+GLGF D GLP+ +P+VD
Sbjct 148 FMGLGFADKKSTRGLPAGLVPVVD 171
> 7296077
Length=418
Score = 28.1 bits (61), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 20/90 (22%), Positives = 37/90 (41%), Gaps = 7/90 (7%)
Query 47 RGGCAPHEKFDPKYSSTFSPIRSLTGDPAVAFIQYGTGACVL-RMGRDIVEIGGIKVPNQ 105
+ C H+ ++ S T+ G P IQ+ + + + D +G + + NQ
Sbjct 126 KQSCLHHDGYNSSESQTYQA----NGSPFQ--IQFASQEILTGILSTDTFTLGDLVIKNQ 179
Query 106 AIGLAVEESTHPFADLPFDGLVGLGFPDVS 135
T FDG++GLGF +++
Sbjct 180 TFAEINSAPTDMCKRSNFDGIIGLGFSEIA 209
> At2g16650
Length=369
Score = 27.7 bits (60), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 18/62 (29%), Positives = 27/62 (43%), Gaps = 4/62 (6%)
Query 42 YLSTHRGGCAPHEKFDPKYSSTFSPIRSLTGDPAVAFIQYGTGACVLRMGRDIVEIGGIK 101
YL+ RG E FD SS SP + A G G ++ ++ V +GG+
Sbjct 87 YLAIDRG----FEIFDRMVSSGISPNEASVTSVARLAAAKGNGDYAFKVVKEFVSVGGVS 142
Query 102 VP 103
+P
Sbjct 143 IP 144
Lambda K H
0.318 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2208717646
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40