bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2325_orf1
Length=130
Score E
Sequences producing significant alignments: (Bits) Value
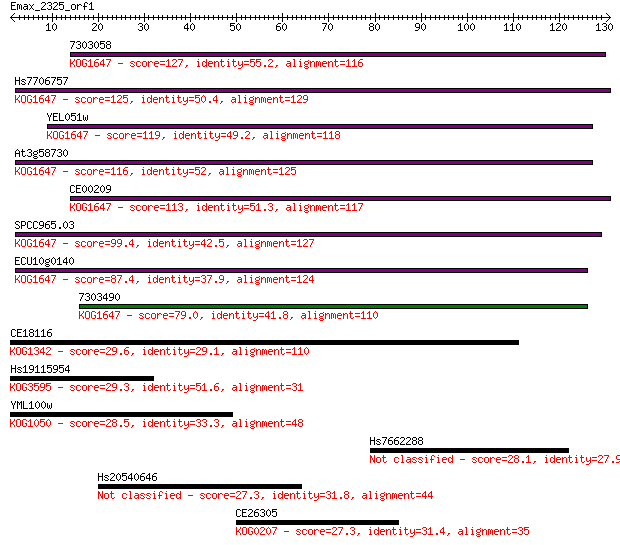
7303058 127 4e-30
Hs7706757 125 3e-29
YEL051w 119 2e-27
At3g58730 116 9e-27
CE00209 113 9e-26
SPCC965.03 99.4 2e-21
ECU10g0140 87.4 6e-18
7303490 79.0 2e-15
CE18116 29.6 1.5
Hs19115954 29.3 1.9
YML100w 28.5 3.7
Hs7662288 28.1 4.5
Hs20540646 27.3 7.4
CE26305 27.3 8.4
> 7303058
Length=246
Score = 127 bits (320), Expect = 4e-30, Method: Compositional matrix adjust.
Identities = 64/116 (55%), Positives = 86/116 (74%), Gaps = 1/116 (0%)
Query 14 DNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAELVKLASLQTAFFT 73
DNVAGV LPVF+ D + D + L+ GG + + YQ A+ LV+LASLQT+F T
Sbjct 99 DNVAGVTLPVFESYQDGS-DTYELAGLARGGQQLAKLKKNYQSAVKLLVELASLQTSFVT 157
Query 74 LDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKKIQEKKKVAR 129
LD IK+TNRRVNA+ +V++P++DR++ YI ELDE+EREEF+RLKKIQ+KK+ AR
Sbjct 158 LDEVIKITNRRVNAIEHVIIPRIDRTLAYIISELDELEREEFYRLKKIQDKKREAR 213
> Hs7706757
Length=247
Score = 125 bits (313), Expect = 3e-29, Method: Compositional matrix adjust.
Identities = 65/129 (50%), Positives = 91/129 (70%), Gaps = 1/129 (0%)
Query 2 IKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAEL 61
+ + V + A DNVAGV LPVF+ + T D + L+ GG + + Y +A+ L
Sbjct 87 VNKAQVKIRAKKDNVAGVTLPVFEHYHEGT-DSYELTGLARGGEQLAKLKRNYAKAVELL 145
Query 62 VKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKKI 121
V+LASLQT+F TLD IK+TNRRVNA+ +V++P+++R++ YI ELDE EREEF+RLKKI
Sbjct 146 VELASLQTSFVTLDEAIKITNRRVNAIEHVIIPRIERTLAYIITELDEREREEFYRLKKI 205
Query 122 QEKKKVARE 130
QEKKK+ +E
Sbjct 206 QEKKKILKE 214
> YEL051w
Length=256
Score = 119 bits (297), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 58/118 (49%), Positives = 82/118 (69%), Gaps = 0/118 (0%)
Query 9 VTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAELVKLASLQ 68
V A +NV+GV L F+ DP ++ + L GG + A++ Y A+ LV+LASLQ
Sbjct 94 VRARQENVSGVYLSQFESYIDPEINDFRLTGLGRGGQQVQRAKEIYSRAVETLVELASLQ 153
Query 69 TAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKKIQEKKK 126
TAF LD IK+TNRRVNA+ +V++P+ + +I YI ELDE++REEF+RLKK+QEKK+
Sbjct 154 TAFIILDEVIKVTNRRVNAIEHVIIPRTENTIAYINSELDELDREEFYRLKKVQEKKQ 211
> At3g58730
Length=261
Score = 116 bits (291), Expect = 9e-27, Method: Compositional matrix adjust.
Identities = 65/126 (51%), Positives = 85/126 (67%), Gaps = 3/126 (2%)
Query 2 IKRPAVFVTAAYDNVAGVRLPVF-QITTDPTVDILKNINLSAGGHVILAARDKYQEALAE 60
+K + V + +N+AGV+LP F + T + L L+ GG + A R Y +A+
Sbjct 89 VKEATLKVRSRTENIAGVKLPKFDHFSEGETKNDL--TGLARGGQQVRACRVAYVKAIEV 146
Query 61 LVKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKK 120
LV+LASLQT+F TLD IK TNRRVNAL NVV PKL+ +I+YI ELDE+ERE+FFRLKK
Sbjct 147 LVELASLQTSFLTLDEAIKTTNRRVNALENVVKPKLENTISYIKGELDELEREDFFRLKK 206
Query 121 IQEKKK 126
IQ K+
Sbjct 207 IQGYKR 212
> CE00209
Length=257
Score = 113 bits (283), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 60/117 (51%), Positives = 78/117 (66%), Gaps = 1/117 (0%)
Query 14 DNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAELVKLASLQTAFFT 73
+NV GV LPVF D D L GG I + Y +A+ LV+LA+LQT F T
Sbjct 101 ENVVGVFLPVFDAYQDGP-DAYDLTGLGKGGANIARLKKNYNKAIELLVELATLQTCFIT 159
Query 74 LDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKKIQEKKKVARE 130
LD IK+TNRRVNA+ +V++P+++ ++TYI ELDEMEREEFFR+KKIQ KK +E
Sbjct 160 LDEAIKVTNRRVNAIEHVIIPRIENTLTYIVTELDEMEREEFFRMKKIQANKKKLKE 216
> SPCC965.03
Length=285
Score = 99.4 bits (246), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 54/127 (42%), Positives = 88/127 (69%), Gaps = 0/127 (0%)
Query 2 IKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAEL 61
+K+P + V + +N++GV LP F++ D ++D + L GG I AR Y++A+ L
Sbjct 88 VKQPRLRVRSKQENISGVFLPTFEMNLDESIDDFQLTGLGKGGQQIQKARQVYEKAVETL 147
Query 62 VKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKKI 121
V+LAS Q+AF L ++MTNRRVN++ ++++P+L+ +I YI EL+E+ERE+F RLKK+
Sbjct 148 VQLASYQSAFVLLGDVLQMTNRRVNSIEHIIIPRLENTIKYIESELEELEREDFTRLKKV 207
Query 122 QEKKKVA 128
Q+ K+ A
Sbjct 208 QKTKENA 214
> ECU10g0140
Length=212
Score = 87.4 bits (215), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 47/124 (37%), Positives = 74/124 (59%), Gaps = 3/124 (2%)
Query 2 IKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAEL 61
++ V+V + + V+GV LP F + + IL L G + R+K+ E L L
Sbjct 85 CQKQNVYVRSRVEQVSGVSLPFFSLQKENIQPILF---LDRSGQSLNECREKFLEVLEML 141
Query 62 VKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKKI 121
V L +L+ +F L++ + TNRRVNAL ++P+L+ +++YI ELDE +R +FFRLKK+
Sbjct 142 VDLCALKNSFRVLNSILMSTNRRVNALEFNIIPRLENTVSYIVSELDEQDRGDFFRLKKV 201
Query 122 QEKK 125
Q K
Sbjct 202 QNLK 205
> 7303490
Length=373
Score = 79.0 bits (193), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 46/110 (41%), Positives = 63/110 (57%), Gaps = 1/110 (0%)
Query 16 VAGVRLPVFQITTDPTVDILKNINLSAGGHVILAARDKYQEALAELVKLASLQTAFFTLD 75
+ GV+L ++ T V LS GG + RD Y +AL LV+ ASL+ L+
Sbjct 101 IVGVKLNTLELETK-GVGAFPLAGLSCGGMQVSRIRDSYTKALKALVEFASLEYQVRMLE 159
Query 76 AEIKMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKKIQEKK 125
A TN RVNAL +VV+P L + YI EL+E ERE+F+RLK+ Q K+
Sbjct 160 AASLQTNMRVNALEHVVIPILQNTYNYICGELEEFEREDFYRLKRSQAKQ 209
> CE18116
Length=465
Score = 29.6 bits (65), Expect = 1.5, Method: Composition-based stats.
Identities = 32/124 (25%), Positives = 58/124 (46%), Gaps = 14/124 (11%)
Query 1 RIKRPAVFVTAAYDNVAGVRLPVFQITT---DPTVDILKNIN-----LSAGGHVILAARD 52
R + AV + D++AG RL VF +TT V+ +K+ N + GG+ I
Sbjct 250 RFQPEAVVLQCGADSLAGDRLGVFNLTTYGHGKCVEYMKSFNVPLLLVGGGGYTIRNVSR 309
Query 53 --KYQEALA---ELVKLASLQTAFFTLDAEIKMTNRRVNALNNVVLPK-LDRSITYITKE 106
Y+ A+A E+ L F + K+ + + AL+N P+ +D++I + +
Sbjct 310 CWLYETAIALNQEVSDDLPLHDYFDYFIPDYKLHIKPLAALSNFNTPEFIDQTIVALLEN 369
Query 107 LDEM 110
L ++
Sbjct 370 LKQL 373
> Hs19115954
Length=4624
Score = 29.3 bits (64), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 16/31 (51%), Positives = 20/31 (64%), Gaps = 1/31 (3%)
Query 1 RIKRPAVFVTAAYDNVAGVRLPVFQITTDPT 31
+IK+P VFVT D VA + VF I TDP+
Sbjct 107 KIKKPKVFVTEGND-VALTGVCVFFIRTDPS 136
> YML100w
Length=1098
Score = 28.5 bits (62), Expect = 3.7, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 26/48 (54%), Gaps = 3/48 (6%)
Query 1 RIKRPAVFVTAAYDNVAGVRLPVFQITTDPTVDILKNINLSAGGHVIL 48
RI P + +D+ + + P+ Q DPT ++LKN+N S H +L
Sbjct 144 RIASP---IQHEHDSGSRIASPIQQQQQDPTTNLLKNVNKSLLVHSLL 188
> Hs7662288
Length=967
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 12/43 (27%), Positives = 27/43 (62%), Gaps = 0/43 (0%)
Query 79 KMTNRRVNALNNVVLPKLDRSITYITKELDEMEREEFFRLKKI 121
++ + ++++ L LD + TKEL+E++ EE +RL+++
Sbjct 627 ELKAKEIDSMQKQRLDWLDAETSRRTKELNELKAEEMYRLQQL 669
> Hs20540646
Length=524
Score = 27.3 bits (59), Expect = 7.4, Method: Composition-based stats.
Identities = 14/45 (31%), Positives = 23/45 (51%), Gaps = 1/45 (2%)
Query 20 RLPVFQITTDPTVDILKNINLSA-GGHVILAARDKYQEALAELVK 63
++P F++ P D+ KN SA GG + D +Q+ LV+
Sbjct 281 KIPEFEVENSPLSDVAKNTESSAFGGSLPAKKSDPFQKEQDHLVE 325
> CE26305
Length=1238
Score = 27.3 bits (59), Expect = 8.4, Method: Composition-based stats.
Identities = 11/35 (31%), Positives = 23/35 (65%), Gaps = 0/35 (0%)
Query 50 ARDKYQEALAELVKLASLQTAFFTLDAEIKMTNRR 84
A+ K EAL++L+ L + + T+D+E ++T+ +
Sbjct 549 AKGKTSEALSKLMSLQAKEATLVTMDSEGRLTSEK 583
Lambda K H
0.320 0.135 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1246445644
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40