bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2419_orf1
Length=112
Score E
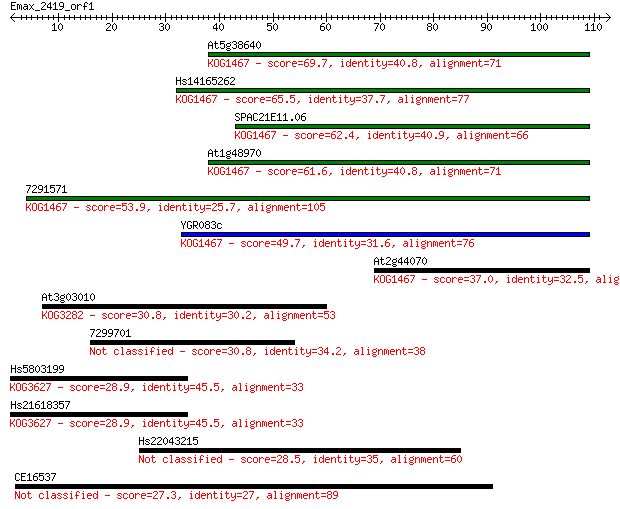
Sequences producing significant alignments: (Bits) Value
At5g38640 69.7 1e-12
Hs14165262 65.5 2e-11
SPAC21E11.06 62.4 2e-10
At1g48970 61.6 4e-10
7291571 53.9 8e-08
YGR083c 49.7 1e-06
At2g44070 37.0 0.009
At3g03010 30.8 0.59
7299701 30.8 0.69
Hs5803199 28.9 2.1
Hs21618357 28.9 2.3
Hs22043215 28.5 3.6
CE16537 27.3 6.4
> At5g38640
Length=642
Score = 69.7 bits (169), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 29/71 (40%), Positives = 46/71 (64%), Gaps = 0/71 (0%)
Query 38 FRQEIIHPAVIRVGLQMANRKIQGANARTVAMLSSFNCFIEDYSPPPYESIDRHIKLSLD 97
F+ + +HPAV +VGLQ + I G NAR +AML +F ++DYS PP +S++R + +
Sbjct 312 FQLDPMHPAVYKVGLQYLSGDISGGNARCIAMLQAFQEVVKDYSTPPEKSLNRDMTAKIS 371
Query 98 RRINFITNCRP 108
++F+ CRP
Sbjct 372 SYVSFLIECRP 382
> Hs14165262
Length=523
Score = 65.5 bits (158), Expect = 2e-11, Method: Composition-based stats.
Identities = 29/77 (37%), Positives = 47/77 (61%), Gaps = 0/77 (0%)
Query 32 LRLLMNFRQEIIHPAVIRVGLQMANRKIQGANARTVAMLSSFNCFIEDYSPPPYESIDRH 91
L M+ +IHPA++R+GLQ + + G+NAR +A+L + I+DY+ PP E + R
Sbjct 192 LTQFMSIPSSVIHPAMVRLGLQYSQGLVSGSNARCIALLRALQQVIQDYTTPPNEELSRD 251
Query 92 IKLSLDRRINFITNCRP 108
+ L ++F+T CRP
Sbjct 252 LVNKLKPYMSFLTQCRP 268
> SPAC21E11.06
Length=467
Score = 62.4 bits (150), Expect = 2e-10, Method: Composition-based stats.
Identities = 27/66 (40%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 43 IHPAVIRVGLQMANRKIQGANARTVAMLSSFNCFIEDYSPPPYESIDRHIKLSLDRRINF 102
IHPAVIR+GL++AN KI G+N R + +L +F I+DY P ++ RH+ ++ +I +
Sbjct 141 IHPAVIRLGLKLANYKIFGSNQRCIDLLKTFKIVIQDYQTPYGTTLSRHLTTHINSQIAY 200
Query 103 ITNCRP 108
+ + RP
Sbjct 201 LVSTRP 206
> At1g48970
Length=756
Score = 61.6 bits (148), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 29/71 (40%), Positives = 41/71 (57%), Gaps = 0/71 (0%)
Query 38 FRQEIIHPAVIRVGLQMANRKIQGANARTVAMLSSFNCFIEDYSPPPYESIDRHIKLSLD 97
F + IH AV +VGLQ I G NAR +AML +F IEDYS PP + + + ++
Sbjct 420 FTLDSIHHAVYKVGLQHLAGDISGDNARCIAMLQAFQEAIEDYSTPPMKDLTMDLTAKIN 479
Query 98 RRINFITNCRP 108
++F+ CRP
Sbjct 480 GYVSFLIECRP 490
> 7291571
Length=626
Score = 53.9 bits (128), Expect = 8e-08, Method: Composition-based stats.
Identities = 27/111 (24%), Positives = 55/111 (49%), Gaps = 6/111 (5%)
Query 4 SSSITASSSTIISSSSSSNSSSSRLPLHLRLLMN------FRQEIIHPAVIRVGLQMANR 57
S + A+SS+ + +S+ + R+ L L+ ++HP++ R+G+Q A R
Sbjct 262 SPTTAATSSSPVKCPASTPNVDCRVKLFNHLVCAKEDSQFINDPLVHPSIARLGVQYAKR 321
Query 58 KIQGANARTVAMLSSFNCFIEDYSPPPYESIDRHIKLSLDRRINFITNCRP 108
+ G+NAR +A L + + D+ P + R + ++ ++ + CRP
Sbjct 322 TVVGSNARCIAFLHALRQVVHDFETPAKKEFGRSLDAAVKHHVDHLHKCRP 372
> YGR083c
Length=651
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 24/76 (31%), Positives = 44/76 (57%), Gaps = 2/76 (2%)
Query 33 RLLMNFRQEIIHPAVIRVGLQMANRKIQGANARTVAMLSSFNCFIEDYSPPPYESIDRHI 92
LL+N +++IHP+++ + +A+ KI G+ R +AML F I+DY P ++ R++
Sbjct 262 ELLLN--KDLIHPSILLLTSHLAHYKIVGSIPRCIAMLEVFQIVIKDYQTPKGTTLSRNL 319
Query 93 KLSLDRRINFITNCRP 108
L +I+ + RP
Sbjct 320 TSYLSHQIDLLKKARP 335
> At2g44070
Length=307
Score = 37.0 bits (84), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 13/40 (32%), Positives = 24/40 (60%), Gaps = 0/40 (0%)
Query 69 MLSSFNCFIEDYSPPPYESIDRHIKLSLDRRINFITNCRP 108
ML +F ++DYS PP +++R + + ++F+ CRP
Sbjct 1 MLQAFQEVVKDYSTPPENTLNRDMTAQISSYVSFLIECRP 40
> At3g03010
Length=179
Score = 30.8 bits (68), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 28/53 (52%), Gaps = 0/53 (0%)
Query 7 ITASSSTIISSSSSSNSSSSRLPLHLRLLMNFRQEIIHPAVIRVGLQMANRKI 59
+++ S I + SS + + S+ PL + L +FR+ V+R L+M KI
Sbjct 31 LSSKSVAIDAGSSGNKKTKSKEPLEIEKLADFRKNFKMVLVVRNDLKMGKGKI 83
> 7299701
Length=638
Score = 30.8 bits (68), Expect = 0.69, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 24/38 (63%), Gaps = 0/38 (0%)
Query 16 SSSSSSNSSSSRLPLHLRLLMNFRQEIIHPAVIRVGLQ 53
S++ + S + RL L + +M R+EI+HP +R+ L+
Sbjct 412 STNPAQQSPNKRLTLREQQVMQLRREIMHPGGVRLNLR 449
> Hs5803199
Length=250
Score = 28.9 bits (63), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 1 TISSSSITASSSTIISSSSSSNSSSSRLPLHLR 33
T+SS +TA +S +IS S++S RLP LR
Sbjct 130 TLSSRCVTAGTSCLISGWGSTSSPQLRLPHTLR 162
> Hs21618357
Length=282
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 1 TISSSSITASSSTIISSSSSSNSSSSRLPLHLR 33
T+SS +TA +S +IS S++S RLP LR
Sbjct 162 TLSSRCVTAGTSCLISGWGSTSSPQLRLPHTLR 194
> Hs22043215
Length=268
Score = 28.5 bits (62), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 21/62 (33%), Positives = 30/62 (48%), Gaps = 7/62 (11%)
Query 25 SSRLPLHLRLLMNFRQEIIHPAVIRV--GLQMANRKIQGANARTVAMLSSFNCFIEDYSP 82
SSRLP LRLL +E++ +R GL NR++ R +A + F+ SP
Sbjct 85 SSRLPPSLRLLPAMTEEVVGDHHLRASKGLSNLNRRV-----RALAPSRAGKAFVATESP 139
Query 83 PP 84
P
Sbjct 140 TP 141
> CE16537
Length=315
Score = 27.3 bits (59), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 24/93 (25%), Positives = 42/93 (45%), Gaps = 6/93 (6%)
Query 2 ISSSSITASSSTIISSSSSSNSSSSRLPLHLRLL----MNFRQEIIHPAVIRVGLQMANR 57
+ S I+ + I SS +N +L RL+ N + ++ AV++ M N
Sbjct 183 VLSKYISVNLEGINISSKQANKFLKKLKEKGRLMSFSATNMKSSLVPSAVVKGTRSMENM 242
Query 58 KIQGANARTVAMLSSFNCFIEDYSPPPYESIDR 90
+I +++ V S FNCF + P Y+ + R
Sbjct 243 EI--SDSAEVERRSVFNCFRPSFVKPEYKFMMR 273
Lambda K H
0.320 0.129 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1188972946
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40