bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2428_orf1
Length=151
Score E
Sequences producing significant alignments: (Bits) Value
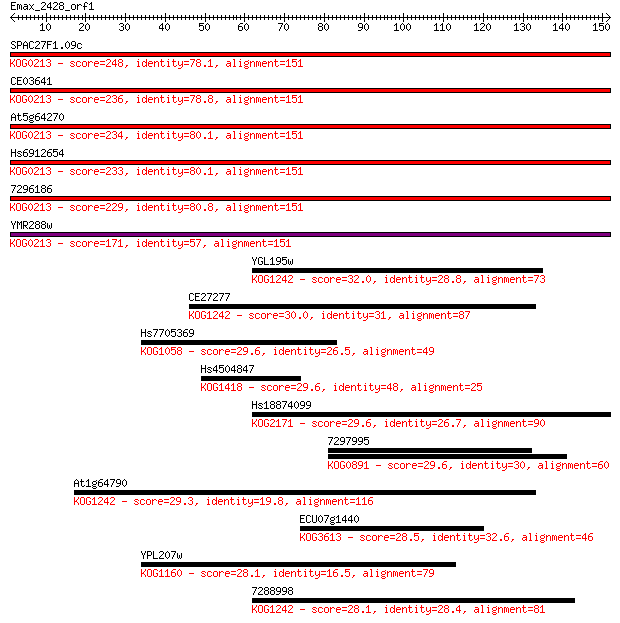
SPAC27F1.09c 248 3e-66
CE03641 236 1e-62
At5g64270 234 3e-62
Hs6912654 233 8e-62
7296186 229 1e-60
YMR288w 171 4e-43
YGL195w 32.0 0.47
CE27277 30.0 1.8
Hs7705369 29.6 2.0
Hs4504847 29.6 2.5
Hs18874099 29.6 2.5
7297995 29.6 2.5
At1g64790 29.3 2.7
ECU07g1440 28.5 5.6
YPL207w 28.1 6.7
7288998 28.1 6.8
> SPAC27F1.09c
Length=1188
Score = 248 bits (633), Expect = 3e-66, Method: Compositional matrix adjust.
Identities = 118/151 (78%), Positives = 134/151 (88%), Gaps = 0/151 (0%)
Query 1 LLIDEDYYARIEGREIISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAFAVVVSALG 60
LLIDEDYYAR EGREIISNLAKA+GLA MIATMRPDIDH DEYVRNTTARAF+VV SALG
Sbjct 465 LLIDEDYYARAEGREIISNLAKASGLAHMIATMRPDIDHVDEYVRNTTARAFSVVASALG 524
Query 61 VPSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLK 120
VP+LL FLKAVC+SKKSWQARHTG+ IIQQIA+LLGC +LPHL+ V+ I HGL+D K
Sbjct 525 VPALLPFLKAVCRSKKSWQARHTGVRIIQQIALLLGCSILPHLKNLVDCIGHGLEDEQQK 584
Query 121 IKTVTALALAALAEAAAPYGIEAFDCVLRPL 151
++ +TAL+L+ALAEAA PYGIEAFD VL+PL
Sbjct 585 VRIMTALSLSALAEAATPYGIEAFDSVLKPL 615
> CE03641
Length=1322
Score = 236 bits (602), Expect = 1e-62, Method: Compositional matrix adjust.
Identities = 119/151 (78%), Positives = 139/151 (92%), Gaps = 0/151 (0%)
Query 1 LLIDEDYYARIEGREIISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAFAVVVSALG 60
LLIDEDYYAR+EGREIISNLAKAAGLATMI+TMRPDID+ DEYVRNTTARAFAVV SALG
Sbjct 599 LLIDEDYYARVEGREIISNLAKAAGLATMISTMRPDIDNVDEYVRNTTARAFAVVASALG 658
Query 61 VPSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLK 120
+P+LL FLKAVC+SKKSWQARHTGI+I+QQ+AIL+GC VLPHL+ V+I++ GLDD K
Sbjct 659 IPALLPFLKAVCKSKKSWQARHTGIKIVQQMAILMGCAVLPHLKALVDIVESGLDDEQQK 718
Query 121 IKTVTALALAALAEAAAPYGIEAFDCVLRPL 151
++T+TAL LAALAEA++PYGIEAFD VL+PL
Sbjct 719 VRTITALCLAALAEASSPYGIEAFDSVLKPL 749
> At5g64270
Length=1269
Score = 234 bits (598), Expect = 3e-62, Method: Compositional matrix adjust.
Identities = 121/151 (80%), Positives = 139/151 (92%), Gaps = 0/151 (0%)
Query 1 LLIDEDYYARIEGREIISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAFAVVVSALG 60
LLIDEDYYAR+EGREIISNL+KAAGLA+MIA MRPDID+ DEYVRNTTARAF+VV SALG
Sbjct 545 LLIDEDYYARVEGREIISNLSKAAGLASMIAAMRPDIDNIDEYVRNTTARAFSVVASALG 604
Query 61 VPSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLK 120
+P+LL FLKAVCQSK+SWQARHTGI+I+QQIAIL+GC VLPHLR VEII+HGL D K
Sbjct 605 IPALLPFLKAVCQSKRSWQARHTGIKIVQQIAILIGCAVLPHLRSLVEIIEHGLSDENQK 664
Query 121 IKTVTALALAALAEAAAPYGIEAFDCVLRPL 151
++T+TAL+LAALAEAAAPYGIE+FD VL+PL
Sbjct 665 VRTITALSLAALAEAAAPYGIESFDSVLKPL 695
> Hs6912654
Length=1304
Score = 233 bits (595), Expect = 8e-62, Method: Compositional matrix adjust.
Identities = 121/151 (80%), Positives = 138/151 (91%), Gaps = 0/151 (0%)
Query 1 LLIDEDYYARIEGREIISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAFAVVVSALG 60
LLIDEDYYAR+EG EIISNLAKAAGLATMI+TMRPDID+ DEYVRNTTARAFAVV SALG
Sbjct 581 LLIDEDYYARVEGLEIISNLAKAAGLATMISTMRPDIDNMDEYVRNTTARAFAVVASALG 640
Query 61 VPSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLK 120
+PSLL FLKAVC+SKKSWQARHTGI+I+QQIAIL+GC +LPHLR VEII+HGL D K
Sbjct 641 IPSLLPFLKAVCKSKKSWQARHTGIKIVQQIAILMGCAILPHLRSLVEIIEHGLVDEQQK 700
Query 121 IKTVTALALAALAEAAAPYGIEAFDCVLRPL 151
++T++ALA+AALAEAA PYGIE+FD VL+PL
Sbjct 701 VRTISALAIAALAEAATPYGIESFDSVLKPL 731
> 7296186
Length=1349
Score = 229 bits (584), Expect = 1e-60, Method: Compositional matrix adjust.
Identities = 122/151 (80%), Positives = 139/151 (92%), Gaps = 0/151 (0%)
Query 1 LLIDEDYYARIEGREIISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAFAVVVSALG 60
LLIDED+YARIEGREIISNLAKAAGLATMI+TMRPDID+ DEYVRNTTARAFAVV SALG
Sbjct 626 LLIDEDHYARIEGREIISNLAKAAGLATMISTMRPDIDNIDEYVRNTTARAFAVVASALG 685
Query 61 VPSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLK 120
+PSLL FLKAVC+SKKSWQARHTGI+I+QQIAIL+GC +LPHL+ VEII+HGL D K
Sbjct 686 IPSLLPFLKAVCKSKKSWQARHTGIKIVQQIAILMGCAILPHLKALVEIIEHGLVDEQQK 745
Query 121 IKTVTALALAALAEAAAPYGIEAFDCVLRPL 151
++T+TALA+AALAEAA PYGIE+FD VL+PL
Sbjct 746 VRTITALAIAALAEAATPYGIESFDSVLKPL 776
> YMR288w
Length=971
Score = 171 bits (433), Expect = 4e-43, Method: Compositional matrix adjust.
Identities = 86/151 (56%), Positives = 107/151 (70%), Gaps = 0/151 (0%)
Query 1 LLIDEDYYARIEGREIISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAFAVVVSALG 60
LLIDED R G+EII+NL+ AGL T++ MRPDI++ DEYVRN T+RA AVV ALG
Sbjct 250 LLIDEDPMVRSTGQEIITNLSTVAGLKTILTVMRPDIENEDEYVRNVTSRAAAVVAKALG 309
Query 61 VPSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLK 120
V LL F+ A C S+KSW+ARHTGI+I+QQI ILLG GVL HL + I+ L D +
Sbjct 310 VNQLLPFINAACHSRKSWKARHTGIKIVQQIGILLGIGVLNHLTGLMSCIKDCLMDDHVP 369
Query 121 IKTVTALALAALAEAAAPYGIEAFDCVLRPL 151
++ VTA L+ LAE + PYGIE F+ VL PL
Sbjct 370 VRIVTAHTLSTLAENSYPYGIEVFNVVLEPL 400
> YGL195w
Length=2672
Score = 32.0 bits (71), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 40/76 (52%), Gaps = 3/76 (3%)
Query 62 PSLLLFLKAVCQS--KKSWQARHTGIEIIQQIAILLGC-GVLPHLRQFVEIIQHGLDDPV 118
PSL L + + + +S + +I+ +AIL+ ++P+L+Q ++ ++ + DPV
Sbjct 1561 PSLALIIHIIHRGMHDRSANIKRKACKIVGNMAILVDTKDLIPYLQQLIDEVEIAMVDPV 1620
Query 119 LKIKTVTALALAALAE 134
+ A AL AL E
Sbjct 1621 PNTRATAARALGALVE 1636
> CE27277
Length=1788
Score = 30.0 bits (66), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 27/95 (28%), Positives = 46/95 (48%), Gaps = 8/95 (8%)
Query 46 NTTARAFAVVVSA-----LGVPSLLLFLKAVCQS--KKSWQARHTGIEIIQQI-AILLGC 97
N T+ A V++ + PSL L + V ++ + + R +II I ++
Sbjct 713 NKTSAALQAVLNTKFIHYIDAPSLALMMPIVRRAFEDRLSETRRVAAQIISNIYSLTENK 772
Query 98 GVLPHLRQFVEIIQHGLDDPVLKIKTVTALALAAL 132
+ P+L V +Q L DPV +I+ V+A AL A+
Sbjct 773 DMEPYLAHMVPGLQRSLLDPVPEIRAVSARALGAV 807
> Hs7705369
Length=953
Score = 29.6 bits (65), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 13/49 (26%), Positives = 27/49 (55%), Gaps = 0/49 (0%)
Query 34 RPDIDHPDEYVRNTTARAFAVVVSALGVPSLLLFLKAVCQSKKSWQARH 82
R D+ HP+E++R +T R + A + L+ ++A + + S+ R+
Sbjct 106 RKDLQHPNEFIRGSTLRFLCKLKEAELLEPLMPAIRACLEHRHSYVRRN 154
> Hs4504847
Length=336
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 17/25 (68%), Gaps = 0/25 (0%)
Query 49 ARAFAVVVSALGVPSLLLFLKAVCQ 73
+AF ++ S +G+P LLFL AV Q
Sbjct 130 GKAFCIIYSVIGIPFTLLFLTAVVQ 154
> Hs18874099
Length=1081
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 45/95 (47%), Gaps = 11/95 (11%)
Query 62 PSLLLFLKAVCQSKKSWQARHTGIEIIQQIAILLGCGVLPHLRQ-----FVEIIQHGLDD 116
P L+ L+ +S+ +Q R G+ ++ +L G H+RQ ++I+ GL+D
Sbjct 351 PQLMPMLEEALRSESPYQ-RKAGLLVL----AVLSDGAGDHIRQRLLPPLLQIVCKGLED 405
Query 117 PVLKIKTVTALALAALAEAAAPYGIEAFDCVLRPL 151
P ++ AL +E P+ I ++ + PL
Sbjct 406 PSQVVRNAALFALGQFSENLQPH-ISSYSREVMPL 439
> 7297995
Length=2470
Score = 29.6 bits (65), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 81 RHTGIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLKIKTVTALALAA 131
RH+ + I++++AI L H+ F E+I + + DP I+ AL A
Sbjct 152 RHSAVFILRELAIALPTYFYQHILTFFEVIFNAIFDPKPAIRESAGEALRA 202
Score = 29.3 bits (64), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 30/60 (50%), Gaps = 0/60 (0%)
Query 81 RHTGIEIIQQIAILLGCGVLPHLRQFVEIIQHGLDDPVLKIKTVTALALAALAEAAAPYG 140
++T I +I+QIA+ LGC +L + + I L K + VT L AL + + G
Sbjct 1023 QNTLINLIEQIAVALGCEFRDYLAELIPQILRVLQHDNSKDRMVTRRLLQALQKFGSTLG 1082
> At1g64790
Length=2428
Score = 29.3 bits (64), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 23/121 (19%), Positives = 62/121 (51%), Gaps = 5/121 (4%)
Query 17 ISNLAKAAGLATMIATMRPDIDHPDEYVRNTTARAF-AVVVSALGVPSLLLFLKAVCQS- 74
+ ++ K +++++ T+ + P+EY R+ V+++ PSL L + V +
Sbjct 1326 VGSVIKNPEISSLVPTLLLALTDPNEYTRHALDTLLQTTFVNSVDAPSLALLVPIVHRGL 1385
Query 75 -KKSWQARHTGIEIIQQIAILLG--CGVLPHLRQFVEIIQHGLDDPVLKIKTVTALALAA 131
++S + + +I+ + L+ ++P++ + ++ L DP+ ++++V A A+ +
Sbjct 1386 RERSSETKKKASQIVGNMCSLVTEPKDMIPYIGLLLPEVKKVLVDPIPEVRSVAARAVGS 1445
Query 132 L 132
L
Sbjct 1446 L 1446
> ECU07g1440
Length=1562
Score = 28.5 bits (62), Expect = 5.6, Method: Composition-based stats.
Identities = 15/53 (28%), Positives = 28/53 (52%), Gaps = 7/53 (13%)
Query 74 SKKSWQARHTGIEIIQQIAILLGCGVLP-------HLRQFVEIIQHGLDDPVL 119
++K + R T +EII+ + IL+ G+L L + V++ + DDP +
Sbjct 879 NRKEYDGRDTDMEIIKILEILVRGGILSGSVMNSVDLSRIVDVAKENRDDPAI 931
> YPL207w
Length=810
Score = 28.1 bits (61), Expect = 6.7, Method: Composition-based stats.
Identities = 13/79 (16%), Positives = 38/79 (48%), Gaps = 11/79 (13%)
Query 34 RPDIDHPDEYVRNTTARAFAVVVSALGVPSLLLFLKAVCQSKKSWQARHTGIEIIQQIAI 93
R ++D P+ + N ++++ GVP ++ + K+++ RH + ++ + +
Sbjct 499 RWEVDEPEYILENALKGHYSMIKQMRGVPGVI-----AERFAKAFEVRHCALSLVGEPIL 553
Query 94 LLGCGVLPHLRQFVEIIQH 112
PH+ +F++++
Sbjct 554 ------YPHINKFIQLLHQ 566
> 7288998
Length=2225
Score = 28.1 bits (61), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 23/84 (27%), Positives = 40/84 (47%), Gaps = 3/84 (3%)
Query 62 PSLLLFLKAVCQS--KKSWQARHTGIEIIQQIAILLGCGVL-PHLRQFVEIIQHGLDDPV 118
PSL L + V ++ +S + R +II + L L P+L + ++ L DPV
Sbjct 1164 PSLALIMPVVQRAFMDRSTETRKMAAQIIGNMYSLTDQKDLAPYLPSIIPGLKSSLLDPV 1223
Query 119 LKIKTVTALALAALAEAAAPYGIE 142
+++ V+A AL A+ + E
Sbjct 1224 PEVRAVSARALGAMVKGMGESSFE 1247
Lambda K H
0.325 0.139 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1888713112
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40