bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2457_orf1
Length=119
Score E
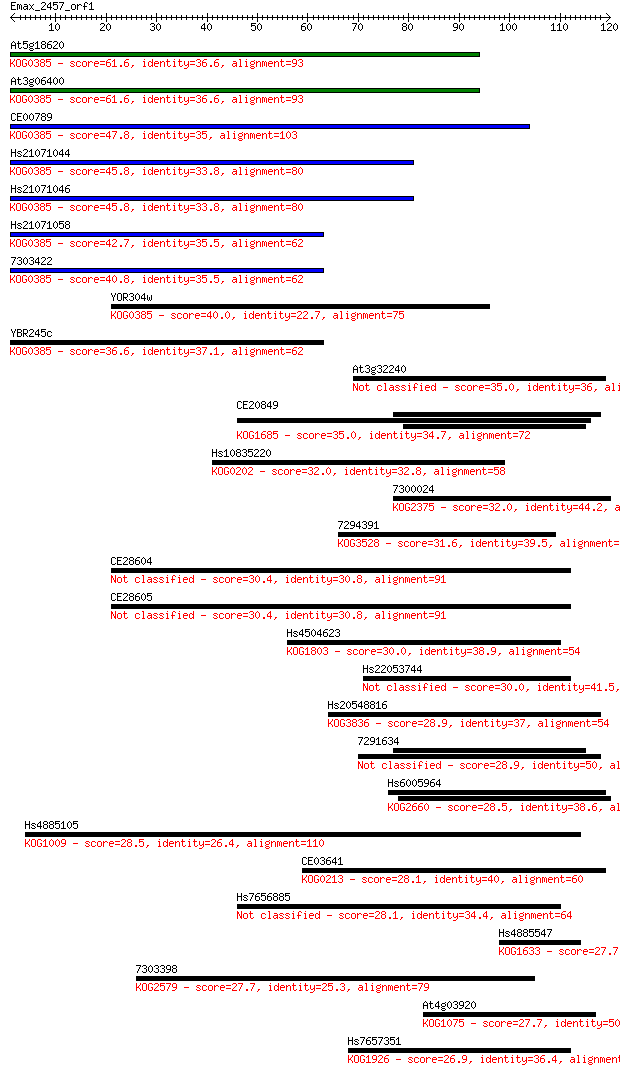
Sequences producing significant alignments: (Bits) Value
At5g18620 61.6 3e-10
At3g06400 61.6 3e-10
CE00789 47.8 5e-06
Hs21071044 45.8 2e-05
Hs21071046 45.8 2e-05
Hs21071058 42.7 2e-04
7303422 40.8 5e-04
YOR304w 40.0 0.001
YBR245c 36.6 0.011
At3g32240 35.0 0.029
CE20849 35.0 0.030
Hs10835220 32.0 0.30
7300024 32.0 0.31
7294391 31.6 0.32
CE28604 30.4 0.73
CE28605 30.4 0.76
Hs4504623 30.0 0.95
Hs22053744 30.0 1.1
Hs20548816 28.9 2.4
7291634 28.9 2.5
Hs6005964 28.5 2.9
Hs4885105 28.5 3.2
CE03641 28.1 3.8
Hs7656885 28.1 4.5
Hs4885547 27.7 4.8
7303398 27.7 4.9
At4g03920 27.7 5.0
Hs7657351 26.9 9.3
> At5g18620
Length=1063
Score = 61.6 bits (148), Expect = 3e-10, Method: Composition-based stats.
Identities = 34/100 (34%), Positives = 56/100 (56%), Gaps = 7/100 (7%)
Query 1 FTEEEDIFVLNLTTLLGYGNWEKLRSQILRDSNWVGDWFFRSRAAADLGRRAEAIIRQLK 60
+ EE D F++ + LGYGNW++L++ + DWF +SR +L RR + +IR ++
Sbjct 943 YNEECDRFMICMVHKLGYGNWDELKAAFRTSPLFRFDWFVKSRTTQELARRCDTLIRLIE 1002
Query 61 KE-----EGERFSRGRRRLDMPATKQQSPSA--ATGTPAA 93
KE E ER +R ++L AT + PS A +P++
Sbjct 1003 KENQEFDERERQARKEKKLSKSATPSKRPSGRQANESPSS 1042
> At3g06400
Length=1057
Score = 61.6 bits (148), Expect = 3e-10, Method: Composition-based stats.
Identities = 34/100 (34%), Positives = 58/100 (58%), Gaps = 7/100 (7%)
Query 1 FTEEEDIFVLNLTTLLGYGNWEKLRSQILRDSNWVGDWFFRSRAAADLGRRAEAIIRQLK 60
+ EE D F++ + LGYGNW++L++ S + DWF +SR + +L RR + +IR ++
Sbjct 948 YNEECDRFMICMIHKLGYGNWDELKAAFRTSSVFRFDWFVKSRTSQELARRCDTLIRLIE 1007
Query 61 KE-----EGERFSRGRRRLDMPATKQQSP--SAATGTPAA 93
KE E ER +R ++L AT + P A+ +P++
Sbjct 1008 KENQEFDERERQARKEKKLAKSATPSKRPLGRQASESPSS 1047
> CE00789
Length=971
Score = 47.8 bits (112), Expect = 5e-06, Method: Composition-based stats.
Identities = 36/110 (32%), Positives = 56/110 (50%), Gaps = 11/110 (10%)
Query 1 FTEEEDIFVLNLTTLLGYGN---WEKLRSQILRDSNWVGDWFFRSRAAADLGRRAEAIIR 57
+TEEED F++ T LG+ +E+LR + + DWF +SR A +L RR +I
Sbjct 865 YTEEEDRFLVCETHRLGHDKENVFEELRQSVRMAPQFRFDWFLKSRTAMELQRRCNTLIT 924
Query 58 QLKKEEGERFSRGRRRLDMPATKQQSP----SAATGTPAAATGTPAAATP 103
+++E GE + + A K++S S ++GTP T ATP
Sbjct 925 LIEREMGE-VVESKPVIVTAADKKKSVAKDLSKSSGTP---TAKKVKATP 970
> Hs21071044
Length=1054
Score = 45.8 bits (107), Expect = 2e-05, Method: Composition-based stats.
Identities = 27/88 (30%), Positives = 44/88 (50%), Gaps = 8/88 (9%)
Query 1 FTEEEDIFVLNLTTLLGYGN---WEKLRSQILRDSNWVGDWFFRSRAAADLGRRAEAIIR 57
+TEEED F++ + +G+ +E+LR + + DWF +SR A + RR +I
Sbjct 962 YTEEEDRFLICMLHKMGFDRENVYEELRQCVRNAPQFRFDWFIKSRTAMEFQRRCNTLIS 1021
Query 58 QLKK-----EEGERFSRGRRRLDMPATK 80
++K EE ER + +R P K
Sbjct 1022 LIEKENMEIEERERAEKKKRATKTPMVK 1049
> Hs21071046
Length=1033
Score = 45.8 bits (107), Expect = 2e-05, Method: Composition-based stats.
Identities = 27/88 (30%), Positives = 44/88 (50%), Gaps = 8/88 (9%)
Query 1 FTEEEDIFVLNLTTLLGYGN---WEKLRSQILRDSNWVGDWFFRSRAAADLGRRAEAIIR 57
+TEEED F++ + +G+ +E+LR + + DWF +SR A + RR +I
Sbjct 941 YTEEEDRFLICMLHKMGFDRENVYEELRQCVRNAPQFRFDWFIKSRTAMEFQRRCNTLIS 1000
Query 58 QLKK-----EEGERFSRGRRRLDMPATK 80
++K EE ER + +R P K
Sbjct 1001 LIEKENMEIEERERAEKKKRATKTPMVK 1028
> Hs21071058
Length=1052
Score = 42.7 bits (99), Expect = 2e-04, Method: Composition-based stats.
Identities = 22/65 (33%), Positives = 37/65 (56%), Gaps = 3/65 (4%)
Query 1 FTEEEDIFVLNLTTLLGYGN---WEKLRSQILRDSNWVGDWFFRSRAAADLGRRAEAIIR 57
+TEEED F++ + LG+ +++LR I + DWF +SR A +L RR +I
Sbjct 947 YTEEEDRFLICMLHKLGFDKENVYDELRQCIRNSPQFRFDWFLKSRTAMELQRRCNTLIT 1006
Query 58 QLKKE 62
+++E
Sbjct 1007 LIERE 1011
> 7303422
Length=1027
Score = 40.8 bits (94), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 37/65 (56%), Gaps = 3/65 (4%)
Query 1 FTEEEDIFVLNLTTLLGYGN---WEKLRSQILRDSNWVGDWFFRSRAAADLGRRAEAIIR 57
+TE ED F++ + LG+ +E+LR+ I + DWF +SR A +L RR +I
Sbjct 902 YTEIEDRFLVCMLHKLGFDKENVYEELRAAIRASPQFRFDWFIKSRTALELQRRCNTLIT 961
Query 58 QLKKE 62
+++E
Sbjct 962 LIERE 966
> YOR304w
Length=1120
Score = 40.0 bits (92), Expect = 0.001, Method: Composition-based stats.
Identities = 17/75 (22%), Positives = 40/75 (53%), Gaps = 0/75 (0%)
Query 21 WEKLRSQILRDSNWVGDWFFRSRAAADLGRRAEAIIRQLKKEEGERFSRGRRRLDMPATK 80
+EKL+ +I+ + DWF ++R +L +R ++ + +E + + ++R AT+
Sbjct 1017 YEKLKQEIMMSDLFTFDWFIKTRTVHELSKRVHTLLTLIVREYEQPDANKKKRSRTSATR 1076
Query 81 QQSPSAATGTPAAAT 95
+ +P + + A+T
Sbjct 1077 EDTPLSQNESTRAST 1091
> YBR245c
Length=1129
Score = 36.6 bits (83), Expect = 0.011, Method: Composition-based stats.
Identities = 23/67 (34%), Positives = 38/67 (56%), Gaps = 7/67 (10%)
Query 1 FTEEEDIFVLNLTTLLGYG-----NWEKLRSQILRDSNWVGDWFFRSRAAADLGRRAEAI 55
++EEED F+L + L YG +E +R +I + D++FRSR +L RR +
Sbjct 992 YSEEEDRFILLM--LFKYGLDRDDVYELVRDEIRDCPLFELDFYFRSRTPVELARRGNTL 1049
Query 56 IRQLKKE 62
++ L+KE
Sbjct 1050 LQCLEKE 1056
> At3g32240
Length=487
Score = 35.0 bits (79), Expect = 0.029, Method: Compositional matrix adjust.
Identities = 18/50 (36%), Positives = 30/50 (60%), Gaps = 0/50 (0%)
Query 69 RGRRRLDMPATKQQSPSAATGTPAAATGTPAAATPAGTTPAAATPAATAA 118
RR PA SP+ +TG+PA +TG+PA + + ++PA +P+ A+
Sbjct 43 HSRRHSGSPAMSTGSPAQSTGSPAQSTGSPALFSGSPSSPATGSPSTPAS 92
> CE20849
Length=543
Score = 35.0 bits (79), Expect = 0.030, Method: Composition-based stats.
Identities = 19/41 (46%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 77 PATKQQSPSAATGTPAAATGTPAAATPAGTTPAAATPAATA 117
P T ++ + TPA T T AA TP TPAA TPA A
Sbjct 58 PKTPSRTTKSTVDTPAPKTPTRAAKTPVVKTPAAKTPAKKA 98
Score = 30.0 bits (66), Expect = 0.94, Method: Composition-based stats.
Identities = 23/76 (30%), Positives = 33/76 (43%), Gaps = 6/76 (7%)
Query 46 ADLGRRAEAIIRQLKKEEGERFSRGRRRLD----MPATKQQSPSAATGTPAAATGTPAAA 101
AD+ + E ++ K +R ++ L PA K +P + T + TPA
Sbjct 16 ADVPEKDEEVVVPPKTPARSPKTRAQKSLAAGGATPAPKTPAPKTPSRTTKSTVDTPAPK 75
Query 102 TP--AGTTPAAATPAA 115
TP A TP TPAA
Sbjct 76 TPTRAAKTPVVKTPAA 91
Score = 29.6 bits (65), Expect = 1.2, Method: Composition-based stats.
Identities = 15/36 (41%), Positives = 16/36 (44%), Gaps = 0/36 (0%)
Query 79 TKQQSPSAATGTPAAATGTPAAATPAGTTPAAATPA 114
TK + A TP A TP TPA TPA P
Sbjct 65 TKSTVDTPAPKTPTRAAKTPVVKTPAAKTPAKKAPG 100
> Hs10835220
Length=994
Score = 32.0 bits (71), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 28/59 (47%), Gaps = 1/59 (1%)
Query 41 RSRAAADLGRRAEAIIRQL-KKEEGERFSRGRRRLDMPATKQQSPSAATGTPAAATGTP 98
RS + + ++IRQL KKE FSR R+ + + + +S AA G G P
Sbjct 460 RSLSKVERANACNSVIRQLMKKEFTLEFSRDRKSMSVYCSPAKSSRAAVGNKMFVKGAP 518
> 7300024
Length=1069
Score = 32.0 bits (71), Expect = 0.31, Method: Composition-based stats.
Identities = 19/48 (39%), Positives = 22/48 (45%), Gaps = 5/48 (10%)
Query 77 PATKQQSPSAATGTPAAATGTPAAATPAG-----TTPAAATPAATAAT 119
P PS+ T TP TP A A TTPAA TP + +AT
Sbjct 773 PEVSPAPPSSNTTTPTGIASTPTAGVIASAGSEKTTPAAPTPTSNSAT 820
> 7294391
Length=1552
Score = 31.6 bits (70), Expect = 0.32, Method: Composition-based stats.
Identities = 17/47 (36%), Positives = 24/47 (51%), Gaps = 4/47 (8%)
Query 66 RFSR----GRRRLDMPATKQQSPSAATGTPAAATGTPAAATPAGTTP 108
+FSR G +L +PA KQQSP + + P +T T P +P
Sbjct 424 KFSRAKTEGEIKLHLPAPKQQSPQSKSRIPIVSTPTAQKTVPKPVSP 470
> CE28604
Length=2257
Score = 30.4 bits (67), Expect = 0.73, Method: Composition-based stats.
Identities = 28/110 (25%), Positives = 43/110 (39%), Gaps = 19/110 (17%)
Query 21 WEKLR-----SQILRDSNWVGDWFFRSRA---AADLGRRAEAIIRQLKKEEG---ERFSR 69
WE L+ Q RD+ W F + + GR E I+ +KK E F++
Sbjct 2000 WEYLQLILEVYQFARDAAVAESWLFAQEPYLISKEYGRNLEETIKLIKKHEAFEKSAFAQ 2059
Query 70 GRRRL--------DMPATKQQSPSAATGTPAAATGTPAAATPAGTTPAAA 111
R L ++ T+ + A A G+P+ +TPA T A
Sbjct 2060 EERFLALEKLTTFELKETQHREEETAKRRGPAHIGSPSRSTPAAETSFGA 2109
> CE28605
Length=2182
Score = 30.4 bits (67), Expect = 0.76, Method: Composition-based stats.
Identities = 28/110 (25%), Positives = 43/110 (39%), Gaps = 19/110 (17%)
Query 21 WEKLR-----SQILRDSNWVGDWFFRSRA---AADLGRRAEAIIRQLKKEEG---ERFSR 69
WE L+ Q RD+ W F + + GR E I+ +KK E F++
Sbjct 2000 WEYLQLILEVYQFARDAAVAESWLFAQEPYLISKEYGRNLEETIKLIKKHEAFEKSAFAQ 2059
Query 70 GRRRL--------DMPATKQQSPSAATGTPAAATGTPAAATPAGTTPAAA 111
R L ++ T+ + A A G+P+ +TPA T A
Sbjct 2060 EERFLALEKLTTFELKETQHREEETAKRRGPAHIGSPSRSTPAAETSFGA 2109
> Hs4504623
Length=993
Score = 30.0 bits (66), Expect = 0.95, Method: Compositional matrix adjust.
Identities = 21/57 (36%), Positives = 27/57 (47%), Gaps = 4/57 (7%)
Query 56 IRQLKKEEGERFS---RGRRRLDMPATKQQSPSAATGTPAAATGTPAAATPAGTTPA 109
+ Q+ +E G R G+RR + + P AA G P A TG PA P TPA
Sbjct 758 VHQIAEEHGLRHDSSGEGKRRFITVSKRAPRPRAALG-PPAGTGGPAPLQPVPPTPA 813
> Hs22053744
Length=679
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 22/41 (53%), Gaps = 0/41 (0%)
Query 71 RRRLDMPATKQQSPSAATGTPAAATGTPAAATPAGTTPAAA 111
R ++D A +QQ S A +PA TPAGTTP A+
Sbjct 188 REQIDFLAARQQFLSLEQANKGAPHSSPARGTPAGTTPGAS 228
> Hs20548816
Length=265
Score = 28.9 bits (63), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 28/56 (50%), Gaps = 2/56 (3%)
Query 64 GERFSRGRRRLDMPATKQQSPSAATGTPAAATGTPAAATPAGTTP--AAATPAATA 117
G ++ G L +P + P ATG P + T T AA +PA P A + P A+A
Sbjct 183 GNKWGIGLDVLLVPTVRGHRPDPATGQPPSVTKTWAARSPASEAPGKAGSAPKASA 238
> 7291634
Length=157
Score = 28.9 bits (63), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 22/41 (53%), Positives = 26/41 (63%), Gaps = 3/41 (7%)
Query 77 PATKQQSPSAATGTPAA---ATGTPAAATPAGTTPAAATPA 114
PA SP ATGTP + ATGTP +A+PA TP + TPA
Sbjct 35 PAGTPTSPPPATGTPPSPSPATGTPPSASPAAGTPTSPTPA 75
Score = 26.9 bits (58), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 24/49 (48%), Positives = 28/49 (57%), Gaps = 3/49 (6%)
Query 70 GRRRLDMPAT-KQQSPSAATGTPAAATGTPAAATPAGTTPAAATPAATA 117
G PAT SPS ATGTP +A+ PAA TP TPA TP++ A
Sbjct 37 GTPTSPPPATGTPPSPSPATGTPPSAS--PAAGTPTSPTPATGTPSSPA 83
> Hs6005964
Length=344
Score = 28.5 bits (62), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 24/43 (55%), Gaps = 4/43 (9%)
Query 76 MPATKQQSPSAATGTPAAATGTPAAATPAGTTPAAATPAATAA 118
+PAT PS AT + G+P++ P T P + TP +TA+
Sbjct 269 LPATSSSLPSPAT----PSHGSPSSHGPPATHPTSPTPPSTAS 307
Score = 26.9 bits (58), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 15/42 (35%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Query 78 ATKQQSPSAATGTPAAATGTPAAATPAGTTPAAATPAATAAT 119
+ ++PS AT PA ++ P+ ATP+ +P++ P AT T
Sbjct 258 SVSDKAPSPAT-LPATSSSLPSPATPSHGSPSSHGPPATHPT 298
> Hs4885105
Length=559
Score = 28.5 bits (62), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 29/136 (21%), Positives = 48/136 (35%), Gaps = 26/136 (19%)
Query 4 EEDIFVLNLTTLLGYGNWEKLRSQILRDSNWVGDWFFRSRAAAD---------------- 47
E+ + + + +G + L D +W D F + ++ D
Sbjct 324 EDSVLLYDTQQSFPFGYVSNIHYHTLSDISWSSDGAFLAISSTDGYCSFVTFEKDELGIP 383
Query 48 ------LGRRAEAIIRQLKKEEGERFSRGRRRLD-MPATKQQSPSAATGTPAAATGTPAA 100
L R ++ K + S G R ++ PA++ Q PS+ TP A PA
Sbjct 384 LKEKPVLNMRTPDTAKKTKSQTHRGSSPGPRPVEGTPASRTQDPSSPGTTPPQARQAPAP 443
Query 101 AT---PAGTTPAAATP 113
P TPA +P
Sbjct 444 TVIRDPPSITPAVKSP 459
> CE03641
Length=1322
Score = 28.1 bits (61), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 24/68 (35%), Positives = 33/68 (48%), Gaps = 10/68 (14%)
Query 59 LKKEEGERFSRGRRRLDMPATKQQSPSAATGTPAAA-------TGTPAAATPAG-TTPAA 110
+K E+ S+ R R D+ T Q+P+ A TP + T + + TP G TP
Sbjct 307 VKIEDTPSASKRRSRWDL--TPSQTPNVAAATPLHSGLQTPSFTPSHPSQTPIGAMTPGG 364
Query 111 ATPAATAA 118
ATP TAA
Sbjct 365 ATPIGTAA 372
> Hs7656885
Length=382
Score = 28.1 bits (61), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 22/67 (32%), Positives = 29/67 (43%), Gaps = 5/67 (7%)
Query 46 ADLGRRAEAIIRQLKKEEGERFSRGRRRLDMPATKQQSPSAATGTPAAATGTPAAATP-- 103
AD G R + R + + R RG+R L + + + P AA A G A A P
Sbjct 25 ADFGARVVRVDRPGSRYDVSRLGRGKRSLVLDLKQPREPRAAASV--QAVGCAAGALPPR 82
Query 104 -AGTTPA 109
G TPA
Sbjct 83 CHGETPA 89
> Hs4885547
Length=1099
Score = 27.7 bits (60), Expect = 4.8, Method: Composition-based stats.
Identities = 11/16 (68%), Positives = 13/16 (81%), Gaps = 0/16 (0%)
Query 98 PAAATPAGTTPAAATP 113
PA+ TPA TTPA+ TP
Sbjct 982 PASTTPASTTPASTTP 997
> 7303398
Length=774
Score = 27.7 bits (60), Expect = 4.9, Method: Composition-based stats.
Identities = 20/82 (24%), Positives = 37/82 (45%), Gaps = 3/82 (3%)
Query 26 SQILRDSNWVGDWFFRSRAAADLGRRAEAIIRQLKKEEGERFSRGRRRL---DMPATKQQ 82
SQ +N+ G W+F A+L R + E + R+ +PA + Q
Sbjct 658 SQTHCAANYEGGWWFSHCQHANLNGRYNLGLTWFDAARNEWIAVKSSRMLVKRLPAVECQ 717
Query 83 SPSAATGTPAAATGTPAAATPA 104
+ ++A+G + +G+ A A P+
Sbjct 718 ANASASGAFVSVSGSAADAAPS 739
> At4g03920
Length=463
Score = 27.7 bits (60), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 17/35 (48%), Positives = 21/35 (60%), Gaps = 1/35 (2%)
Query 83 SPSAATGTPAAATGTPAAATPA-GTTPAAATPAAT 116
+PSAAT PA A P TP+ G +P+ AT AT
Sbjct 90 APSAATSVPAVAPSVPCEVTPSRGPSPSPATAPAT 124
> Hs7657351
Length=1328
Score = 26.9 bits (58), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 25/44 (56%), Gaps = 2/44 (4%)
Query 68 SRGRRRLDMPATKQQSPSAATGTPAAATGTPAAATPAGTTPAAA 111
S GR++ + TK + P+ A GTP + P A T + +TPA +
Sbjct 1207 SMGRKKRN--RTKAKVPAQANGTPTTKSPAPGAPTRSPSTPAKS 1248
Lambda K H
0.314 0.127 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1164469306
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40