bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2510_orf1
Length=83
Score E
Sequences producing significant alignments: (Bits) Value
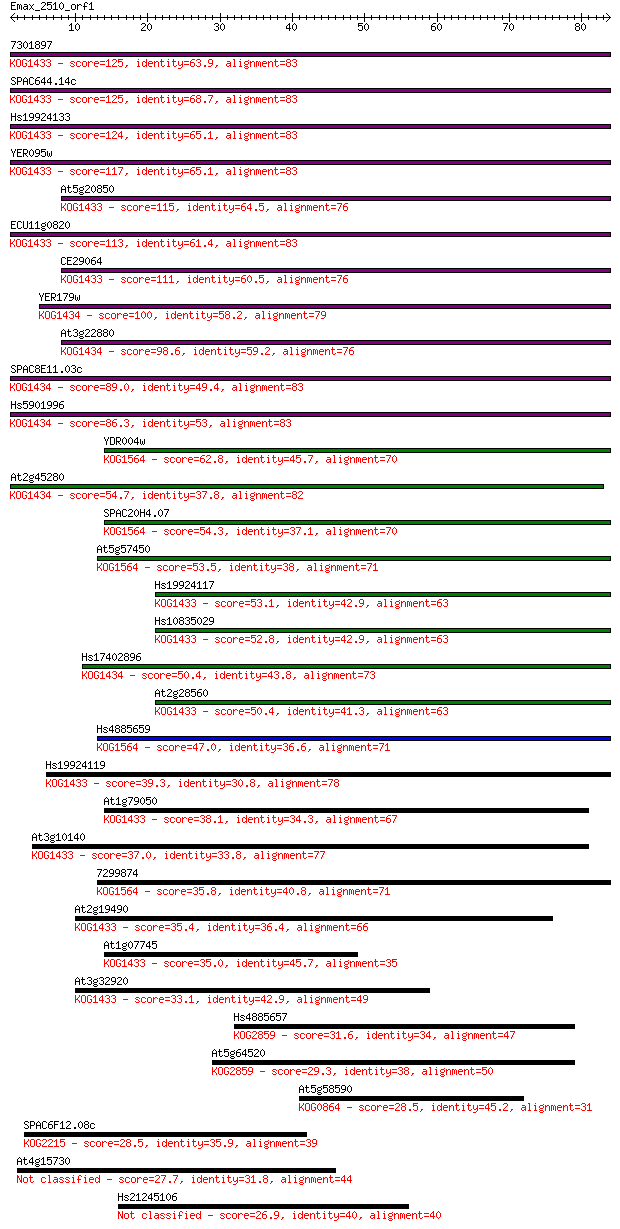
7301897 125 2e-29
SPAC644.14c 125 2e-29
Hs19924133 124 3e-29
YER095w 117 7e-27
At5g20850 115 2e-26
ECU11g0820 113 7e-26
CE29064 111 3e-25
YER179w 100 1e-21
At3g22880 98.6 2e-21
SPAC8E11.03c 89.0 2e-18
Hs5901996 86.3 1e-17
YDR004w 62.8 1e-10
At2g45280 54.7 5e-08
SPAC20H4.07 54.3 5e-08
At5g57450 53.5 1e-07
Hs19924117 53.1 1e-07
Hs10835029 52.8 1e-07
Hs17402896 50.4 7e-07
At2g28560 50.4 9e-07
Hs4885659 47.0 9e-06
Hs19924119 39.3 0.002
At1g79050 38.1 0.005
At3g10140 37.0 0.010
7299874 35.8 0.019
At2g19490 35.4 0.026
At1g07745 35.0 0.031
At3g32920 33.1 0.13
Hs4885657 31.6 0.42
At5g64520 29.3 2.0
At5g58590 28.5 2.8
SPAC6F12.08c 28.5 3.1
At4g15730 27.7 6.0
Hs21245106 26.9 9.0
> 7301897
Length=336
Score = 125 bits (314), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 53/83 (63%), Positives = 71/83 (85%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A+ + + RA++++ +TGS +LD LL GG+ETG++TE+FGEFR GKTQLCHT AV+CQLP+
Sbjct 86 ARTFYQMRADVVQLSTGSKELDKLLGGGIETGSITEIFGEFRCGKTQLCHTLAVTCQLPI 145
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
Q GGEGKC++IDTE TFRPER+
Sbjct 146 SQKGGEGKCMYIDTENTFRPERL 168
> SPAC644.14c
Length=365
Score = 125 bits (313), Expect = 2e-29, Method: Composition-based stats.
Identities = 57/83 (68%), Positives = 69/83 (83%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A EY R+ +I TTGS QLD LL+GGVETG++TELFGEFR GK+Q+CHT AV+CQLP+
Sbjct 111 ATEYHIRRSELITITTGSKQLDTLLQGGVETGSITELFGEFRTGKSQICHTLAVTCQLPI 170
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
+ GGEGKCL+IDTEGTFRP R+
Sbjct 171 DMGGGEGKCLYIDTEGTFRPVRL 193
> Hs19924133
Length=339
Score = 124 bits (312), Expect = 3e-29, Method: Composition-based stats.
Identities = 54/83 (65%), Positives = 72/83 (86%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A E+ + R+ II+ TTGS +LD LL+GG+ETG++TE+FGEFR GKTQ+CHT AV+CQLP+
Sbjct 89 ATEFHQRRSEIIQITTGSKELDKLLQGGIETGSITEMFGEFRTGKTQICHTLAVTCQLPI 148
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
++ GGEGK ++IDTEGTFRPER+
Sbjct 149 DRGGGEGKAMYIDTEGTFRPERL 171
> YER095w
Length=400
Score = 117 bits (292), Expect = 7e-27, Method: Compositional matrix adjust.
Identities = 54/83 (65%), Positives = 67/83 (80%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A ++ R+ +I TTGS LD LL GGVETG++TELFGEFR GK+QLCHT AV+CQ+P+
Sbjct 147 AADFHMRRSELICLTTGSKNLDTLLGGGVETGSITELFGEFRTGKSQLCHTLAVTCQIPL 206
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
+ GGEGKCL+IDTEGTFRP R+
Sbjct 207 DIGGGEGKCLYIDTEGTFRPVRL 229
> At5g20850
Length=339
Score = 115 bits (288), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 49/76 (64%), Positives = 66/76 (86%), Gaps = 0/76 (0%)
Query 8 RANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEG 67
R II+ T+GS +LD +L+GG+ETG++TEL+GEFR+GKTQLCHT V+CQLP++Q GGEG
Sbjct 96 RQEIIQITSGSRELDKVLEGGIETGSITELYGEFRSGKTQLCHTLCVTCQLPMDQGGGEG 155
Query 68 KCLWIDTEGTFRPERV 83
K ++ID EGTFRP+R+
Sbjct 156 KAMYIDAEGTFRPQRL 171
> ECU11g0820
Length=334
Score = 113 bits (283), Expect = 7e-26, Method: Composition-based stats.
Identities = 51/83 (61%), Positives = 65/83 (78%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A Y + R+ ++ TTGS ++D LL GG E+G++TE+FGEFR GKTQLCHT AV+CQLP
Sbjct 83 ASAYHQRRSELVYLTTGSSEVDKLLSGGFESGSITEIFGEFRTGKTQLCHTVAVTCQLPP 142
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
EQ GG GK ++IDTEGTFR ER+
Sbjct 143 EQGGGGGKAMYIDTEGTFRSERL 165
> CE29064
Length=395
Score = 111 bits (277), Expect = 3e-25, Method: Composition-based stats.
Identities = 46/76 (60%), Positives = 62/76 (81%), Gaps = 0/76 (0%)
Query 8 RANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEG 67
R+ +++ TGS LD LL GG+ETG++TE++GE+R GKTQLCH+ AV CQLP++ GGEG
Sbjct 150 RSQLVQIRTGSASLDRLLGGGIETGSITEVYGEYRTGKTQLCHSLAVLCQLPIDMGGGEG 209
Query 68 KCLWIDTEGTFRPERV 83
KC++IDT TFRPER+
Sbjct 210 KCMYIDTNATFRPERI 225
> YER179w
Length=334
Score = 100 bits (248), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 46/79 (58%), Positives = 59/79 (74%), Gaps = 0/79 (0%)
Query 5 LEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSG 64
L+ R + +TGS QLD++L GG+ T ++TE+FGEFR GKTQ+ HT V+ QLP E G
Sbjct 87 LDIRQRVYSLSTGSKQLDSILGGGIMTMSITEVFGEFRCGKTQMSHTLCVTTQLPREMGG 146
Query 65 GEGKCLWIDTEGTFRPERV 83
GEGK +IDTEGTFRPER+
Sbjct 147 GEGKVAYIDTEGTFRPERI 165
> At3g22880
Length=339
Score = 98.6 bits (244), Expect = 2e-21, Method: Composition-based stats.
Identities = 45/76 (59%), Positives = 55/76 (72%), Gaps = 0/76 (0%)
Query 8 RANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEG 67
R +++K TTG LD LL GG+ET +TE FGEFR+GKTQL HT V+ QLP GG G
Sbjct 102 RKSVVKITTGCQALDDLLGGGIETSAITEAFGEFRSGKTQLAHTLCVTTQLPTNMKGGNG 161
Query 68 KCLWIDTEGTFRPERV 83
K +IDTEGTFRP+R+
Sbjct 162 KVAYIDTEGTFRPDRI 177
> SPAC8E11.03c
Length=332
Score = 89.0 bits (219), Expect = 2e-18, Method: Composition-based stats.
Identities = 41/83 (49%), Positives = 58/83 (69%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A E + R + +TGS L+ +L GG+++ ++TE+FGEFR GKTQ+ HT V+ QLP
Sbjct 82 AMEISQNRKKVWSISTGSEALNGILGGGIQSMSITEVFGEFRCGKTQMSHTLCVTAQLPR 141
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
+ G EGK +IDTEGTFRP+R+
Sbjct 142 DMGGAEGKVAFIDTEGTFRPDRI 164
> Hs5901996
Length=340
Score = 86.3 bits (212), Expect = 1e-17, Method: Composition-based stats.
Identities = 44/83 (53%), Positives = 54/83 (65%), Gaps = 0/83 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A EY E R + TTGS + D LL GG+E+ +TE FGEFR GKTQL HT V+ QLP
Sbjct 88 AFEYSEKRKMVFHITTGSQEFDKLLGGGIESMAITEAFGEFRTGKTQLSHTLCVTAQLPG 147
Query 61 EQSGGEGKCLWIDTEGTFRPERV 83
GK ++IDTE TFRP+R+
Sbjct 148 AGGYPGGKIIFIDTENTFRPDRL 170
> YDR004w
Length=460
Score = 62.8 bits (151), Expect = 1e-10, Method: Composition-based stats.
Identities = 32/70 (45%), Positives = 42/70 (60%), Gaps = 0/70 (0%)
Query 14 FTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWID 73
FTT V +D LL GG+ T +TE+FGE GK+QL A+S QL G GKC++I
Sbjct 100 FTTADVAMDELLGGGIFTHGITEIFGESSTGKSQLLMQLALSVQLSEPAGGLGGKCVYIT 159
Query 74 TEGTFRPERV 83
TEG +R+
Sbjct 160 TEGDLPTQRL 169
> At2g45280
Length=332
Score = 54.7 bits (130), Expect = 5e-08, Method: Composition-based stats.
Identities = 31/82 (37%), Positives = 47/82 (57%), Gaps = 0/82 (0%)
Query 1 AQEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPV 60
A + L ++ + TT LD +L GG+ ++TE+ G GKTQ+ +V+ Q+P
Sbjct 62 AWDMLHEEESLPRITTSCSDLDNILGGGISCRDVTEIGGVPGIGKTQIGIQLSVNVQIPR 121
Query 61 EQSGGEGKCLWIDTEGTFRPER 82
E G GK ++IDTEG+F ER
Sbjct 122 ECGGLGGKAIYIDTEGSFMVER 143
> SPAC20H4.07
Length=354
Score = 54.3 bits (129), Expect = 5e-08, Method: Composition-based stats.
Identities = 26/70 (37%), Positives = 38/70 (54%), Gaps = 0/70 (0%)
Query 14 FTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWID 73
TTG V+LD L GG+ G +TE+ GE +GK+Q C + QLP+ G ++I
Sbjct 75 LTTGDVKLDETLHGGIPVGQLTEICGESGSGKSQFCMQLCLMVQLPLSLGGMNKAAVFIS 134
Query 74 TEGTFRPERV 83
TE +R+
Sbjct 135 TESGLETKRL 144
> At5g57450
Length=304
Score = 53.5 bits (127), Expect = 1e-07, Method: Composition-based stats.
Identities = 27/71 (38%), Positives = 39/71 (54%), Gaps = 0/71 (0%)
Query 13 KFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWI 72
K TTG LD L+GG+ ++TE+ E GKTQLC ++ QLP+ G G L++
Sbjct 20 KLTTGCEILDGCLRGGISCDSLTEIVAESGCGKTQLCLQLSLCTQLPISHGGLNGSSLYL 79
Query 73 DTEGTFRPERV 83
+E F R+
Sbjct 80 HSEFPFPFRRL 90
> Hs19924117
Length=350
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 27/63 (42%), Positives = 35/63 (55%), Gaps = 0/63 (0%)
Query 21 LDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWIDTEGTFRP 80
LD L GGV G++TE+ G GKTQ C ++ LP G EG ++IDTE F
Sbjct 90 LDEALHGGVACGSLTEITGPPGCGKTQFCIMMSILATLPTNMGGLEGAVVYIDTESAFSA 149
Query 81 ERV 83
ER+
Sbjct 150 ERL 152
> Hs10835029
Length=350
Score = 52.8 bits (125), Expect = 1e-07, Method: Composition-based stats.
Identities = 27/63 (42%), Positives = 35/63 (55%), Gaps = 0/63 (0%)
Query 21 LDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWIDTEGTFRP 80
LD L GGV G++TE+ G GKTQ C ++ LP G EG ++IDTE F
Sbjct 90 LDEALHGGVACGSLTEITGPPGCGKTQFCIMMSILATLPTNMGGLEGAVVYIDTESAFSA 149
Query 81 ERV 83
ER+
Sbjct 150 ERL 152
> Hs17402896
Length=376
Score = 50.4 bits (119), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 32/73 (43%), Positives = 41/73 (56%), Gaps = 3/73 (4%)
Query 11 IIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCL 70
II F + LD +L GGV TE+ G GKTQLC AV Q+P G G+ +
Sbjct 100 IITFCSA---LDDILGGGVPLMKTTEICGAPGVGKTQLCMQLAVDVQIPECFGGVAGEAV 156
Query 71 WIDTEGTFRPERV 83
+IDTEG+F +RV
Sbjct 157 FIDTEGSFMVDRV 169
> At2g28560
Length=353
Score = 50.4 bits (119), Expect = 9e-07, Method: Composition-based stats.
Identities = 26/63 (41%), Positives = 34/63 (53%), Gaps = 0/63 (0%)
Query 21 LDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWIDTEGTFRP 80
LD L GG+ G +TEL G GK+Q C A+S PV G +G+ ++ID E F
Sbjct 92 LDDTLCGGIPFGVLTELVGPPGIGKSQFCMKLALSASFPVAYGGLDGRVIYIDVESKFSS 151
Query 81 ERV 83
RV
Sbjct 152 RRV 154
> Hs4885659
Length=346
Score = 47.0 bits (110), Expect = 9e-06, Method: Composition-based stats.
Identities = 26/71 (36%), Positives = 38/71 (53%), Gaps = 0/71 (0%)
Query 13 KFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWI 72
+ + G LDALL+GG+ +TEL G GKTQL ++ Q P + G E ++I
Sbjct 81 RLSLGCPVLDALLRGGLPLDGITELAGRSSAGKTQLALQLCLAVQFPRQHGGLEAGAVYI 140
Query 73 DTEGTFRPERV 83
TE F +R+
Sbjct 141 CTEDAFPHKRL 151
> Hs19924119
Length=328
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 24/78 (30%), Positives = 40/78 (51%), Gaps = 5/78 (6%)
Query 6 EARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGG 65
E + + +TG LD LL G+ TG +TE+ G +GKTQ+C A + ++Q+
Sbjct 74 ELKTSTAILSTGIGSLDKLLDAGLYTGEVTEIVGGPGSGKTQVCLCMAANVAHGLQQN-- 131
Query 66 EGKCLWIDTEGTFRPERV 83
L++D+ G R+
Sbjct 132 ---VLYVDSNGGLTASRL 146
> At1g79050
Length=432
Score = 38.1 bits (87), Expect = 0.005, Method: Composition-based stats.
Identities = 23/67 (34%), Positives = 37/67 (55%), Gaps = 6/67 (8%)
Query 14 FTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWID 73
F++G + LD L GG+ G + E++G +GKT L HA++ V++ GG + +D
Sbjct 117 FSSGILTLDLALGGGLPKGRVVEIYGPESSGKTTLA-LHAIA---EVQKLGGNA--MLVD 170
Query 74 TEGTFRP 80
E F P
Sbjct 171 AEHAFDP 177
> At3g10140
Length=389
Score = 37.0 bits (84), Expect = 0.010, Method: Composition-based stats.
Identities = 26/78 (33%), Positives = 40/78 (51%), Gaps = 9/78 (11%)
Query 4 YLEARANIIKFTTGSVQLD-ALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQ 62
Y + R ++I +TGS+ LD AL GG+ G M E++G+ +GKT L + + E
Sbjct 89 YRKRRVSVI--STGSLNLDLALGVGGLPKGRMVEVYGKEASGKTTL------ALHIIKEA 140
Query 63 SGGEGKCLWIDTEGTFRP 80
G C ++D E P
Sbjct 141 QKLGGYCAYLDAENAMDP 158
> 7299874
Length=341
Score = 35.8 bits (81), Expect = 0.019, Method: Compositional matrix adjust.
Identities = 29/72 (40%), Positives = 37/72 (51%), Gaps = 2/72 (2%)
Query 13 KFTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGK-CLW 71
+ + G LD GGV T +TEL G GKTQL ++ QLP E GG GK +
Sbjct 87 RVSFGCSALDRCTGGGVVTRGITELCGAAGVGKTQLLLQLSLCVQLPREL-GGLGKGVAY 145
Query 72 IDTEGTFRPERV 83
I TE +F R+
Sbjct 146 ICTESSFPARRL 157
> At2g19490
Length=376
Score = 35.4 bits (80), Expect = 0.026, Method: Composition-based stats.
Identities = 24/67 (35%), Positives = 36/67 (53%), Gaps = 7/67 (10%)
Query 10 NIIKFTTGSVQLD-ALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGK 68
N+ F+TGS LD AL GG+ G + E++G +GKT L H ++ E G
Sbjct 35 NVPVFSTGSFALDVALGVGGLPKGRVVEIYGPEASGKTTLA-LHVIA-----EAQKQGGT 88
Query 69 CLWIDTE 75
C+++D E
Sbjct 89 CVFVDAE 95
> At1g07745
Length=200
Score = 35.0 bits (79), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 16/35 (45%), Positives = 23/35 (65%), Gaps = 0/35 (0%)
Query 14 FTTGSVQLDALLKGGVETGNMTELFGEFRNGKTQL 48
+TG + D+LL+GG G +TEL G +GKTQ+
Sbjct 67 LSTGDKETDSLLQGGFREGQLTELVGPSSSGKTQM 101
> At3g32920
Length=229
Score = 33.1 bits (74), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 21/50 (42%), Positives = 29/50 (58%), Gaps = 2/50 (4%)
Query 10 NIIKFTTGSVQLD-ALLKGGVETGNMTELFGEFRNGKTQLCHTHAVSCQL 58
N+ F+TGS LD AL GG+ G + E++G +GKT L H +S L
Sbjct 11 NVPVFSTGSFALDVALGVGGLPKGRLVEIYGPEASGKTALA-LHMLSMLL 59
> Hs4885657
Length=280
Score = 31.6 bits (70), Expect = 0.42, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 0/47 (0%)
Query 32 GNMTELFGEFRNGKTQLCHTHAVSCQLPVEQSGGEGKCLWIDTEGTF 78
G++ E G GKT++ + C LP + G E + L+IDT+ F
Sbjct 41 GDILEFHGPEGTGKTEMLYHLTARCILPKSEGGLEVEVLFIDTDYHF 87
> At5g64520
Length=403
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 19/55 (34%), Positives = 28/55 (50%), Gaps = 5/55 (9%)
Query 29 VETGNMTELFGEFRNGKTQLCHTHAVSCQLPVE----QSGGEGK-CLWIDTEGTF 78
+ GN+ E+ G + KTQ+ A+SC LP GG GK L++D + F
Sbjct 40 LRAGNVVEITGASTSAKTQILIQAAISCILPKTWNGIHYGGLGKLVLFLDLDCRF 94
> At5g58590
Length=260
Score = 28.5 bits (62), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 18/33 (54%), Gaps = 2/33 (6%)
Query 41 FRNGKT-QLCHTHAVSCQLPV-EQSGGEGKCLW 71
R KT ++C H +S + V E SG E CLW
Sbjct 101 MRQSKTLKICANHLISSGMSVQEHSGNEKSCLW 133
> SPAC6F12.08c
Length=578
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 14/39 (35%), Positives = 25/39 (64%), Gaps = 1/39 (2%)
Query 3 EYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEF 41
++L+ + ++ +FT L AL++ +E GN+ ELF EF
Sbjct 370 DHLDIQISLQEFTDAIRNL-ALIQNKLERGNLNELFAEF 407
> At4g15730
Length=1052
Score = 27.7 bits (60), Expect = 6.0, Method: Composition-based stats.
Identities = 14/44 (31%), Positives = 25/44 (56%), Gaps = 5/44 (11%)
Query 2 QEYLEARANIIKFTTGSVQLDALLKGGVETGNMTELFGEFRNGK 45
QE+L A+ F+ GS Q+ G+ET N +++ G ++G+
Sbjct 773 QEHLNGDASCRYFSGGSGQIS-----GIETSNSSKVLGSHKSGR 811
> Hs21245106
Length=451
Score = 26.9 bits (58), Expect = 9.0, Method: Composition-based stats.
Identities = 16/42 (38%), Positives = 24/42 (57%), Gaps = 2/42 (4%)
Query 16 TGS--VQLDALLKGGVETGNMTELFGEFRNGKTQLCHTHAVS 55
TGS V +D L+KG + +M + ++ G T L +THA S
Sbjct 81 TGSPPVWMDDLVKGLLSVEDMNVVVVDWNRGATTLIYTHASS 122
Lambda K H
0.317 0.135 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1200681374
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40