bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2550_orf2
Length=111
Score E
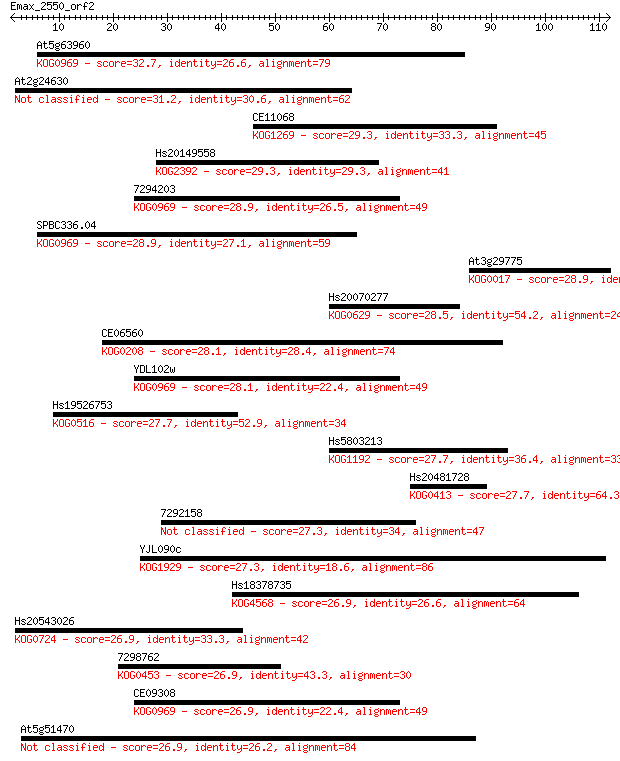
Sequences producing significant alignments: (Bits) Value
At5g63960 32.7 0.18
At2g24630 31.2 0.47
CE11068 29.3 1.9
Hs20149558 29.3 2.0
7294203 28.9 2.2
SPBC336.04 28.9 2.3
At3g29775 28.9 2.6
Hs20070277 28.5 3.3
CE06560 28.1 3.8
YDL102w 28.1 3.8
Hs19526753 27.7 4.8
Hs5803213 27.7 5.0
Hs20481728 27.7 5.3
7292158 27.3 7.7
YJL090c 27.3 7.8
Hs18378735 26.9 8.3
Hs20543026 26.9 9.0
7298762 26.9 9.8
CE09308 26.9 10.0
At5g51470 26.9 10.0
> At5g63960
Length=1081
Score = 32.7 bits (73), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 21/88 (23%), Positives = 42/88 (47%), Gaps = 9/88 (10%)
Query 6 IQLDKPVAESSLDDLRSG-IAKHKEDIQEQILKASKNSELPEAREVKEYAMKEIFVPQTL 64
I D ++ SL+ + + +++ KED+ I+ +N R + Y +K+ ++PQ L
Sbjct 457 IHRDHKLSSYSLNSVSAHFLSEQKEDVHHSIITDLQNGNAETRRRLAVYCLKDAYLPQRL 516
Query 65 -------EEIIEIANKDGV-VPFTVSAG 84
+E+A GV + F ++ G
Sbjct 517 LDKLMFIYNYVEMARVTGVPISFLLARG 544
> At2g24630
Length=690
Score = 31.2 bits (69), Expect = 0.47, Method: Composition-based stats.
Identities = 19/62 (30%), Positives = 31/62 (50%), Gaps = 0/62 (0%)
Query 2 TSFTIQLDKPVAESSLDDLRSGIAKHKEDIQEQILKASKNSELPEAREVKEYAMKEIFVP 61
+S+ + K SS DL + K E + QIL+ +SEL E +V+E + + V
Sbjct 576 SSYEWIVTKKAGRSSESDLLALTDKESEKMPNQILRGVSDSELLEISQVEEQKKQPVSVK 635
Query 62 QT 63
+T
Sbjct 636 KT 637
> CE11068
Length=437
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 23/45 (51%), Gaps = 0/45 (0%)
Query 46 EAREVKEYAMKEIFVPQTLEEIIEIANKDGVVPFTVSAGTPEKKE 90
++ + K Y + + + L+EI +IANK G V TP KE
Sbjct 336 QSDKFKTYVAQRAYFLKNLKEIADIANKTGFVNVQTENMTPRFKE 380
> Hs20149558
Length=376
Score = 29.3 bits (64), Expect = 2.0, Method: Composition-based stats.
Identities = 12/41 (29%), Positives = 23/41 (56%), Gaps = 0/41 (0%)
Query 28 KEDIQEQILKASKNSELPEAREVKEYAMKEIFVPQTLEEII 68
KE+ +E++ K SKN E P K+ K+ ++ + +I+
Sbjct 179 KENTEERLFKVSKNEEKPVQMMFKQSTFKKTYIGEIFTQIL 219
> 7294203
Length=1092
Score = 28.9 bits (63), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 13/50 (26%), Positives = 28/50 (56%), Gaps = 1/50 (2%)
Query 24 IAKHKEDIQEQILKASKNSELPEAREVKEYAMKEIFVP-QTLEEIIEIAN 72
+ + KED+ I+ +N + R + Y +K+ ++P + LE+++ I N
Sbjct 466 LQEQKEDVHHSIITDLQNGDEQTRRRLAMYCLKDAYLPLRLLEKLMAIVN 515
> SPBC336.04
Length=1086
Score = 28.9 bits (63), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 16/60 (26%), Positives = 30/60 (50%), Gaps = 1/60 (1%)
Query 6 IQLDKPVAESSLDDLRSG-IAKHKEDIQEQILKASKNSELPEAREVKEYAMKEIFVPQTL 64
+Q D + SL+ + S + + KED+ I+ +N R + Y +K+ ++PQ L
Sbjct 447 MQRDFKLRSYSLNAVCSQFLGEQKEDVHYSIITDLQNGTADSRRRLAIYCLKDAYLPQRL 506
> At3g29775
Length=1348
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 12/26 (46%), Positives = 16/26 (61%), Gaps = 0/26 (0%)
Query 86 PEKKEEKPVVPVPIPRTRKPTADHVK 111
P K EK V P+ I +KP+ DH+K
Sbjct 660 PSKSLEKGVTPMEIWSGKKPSVDHLK 685
> Hs20070277
Length=493
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 13/24 (54%), Positives = 16/24 (66%), Gaps = 0/24 (0%)
Query 60 VPQTLEEIIEIANKDGVVPFTVSA 83
VP+ LE I +A +G VPF VSA
Sbjct 220 VPEDLERQIGMAEAEGAVPFLVSA 243
> CE06560
Length=1187
Score = 28.1 bits (61), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 21/76 (27%), Positives = 38/76 (50%), Gaps = 5/76 (6%)
Query 18 DDLRSGIAKHKEDIQEQILKASKNSELPEA--REVKEYAMKEIFVPQTLEEIIEIANKDG 75
D+L +G++ +E I++ SK + L E E+ EY +IFV Q++ E+ D
Sbjct 721 DNLLTGLSVAREC---GIIRPSKRAFLVEHVPGELDEYGRTKIFVKQSVSSSDEVIEDDA 777
Query 76 VVPFTVSAGTPEKKEE 91
V ++ + T + E
Sbjct 778 SVSISMCSSTWKGSSE 793
> YDL102w
Length=1097
Score = 28.1 bits (61), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 11/50 (22%), Positives = 28/50 (56%), Gaps = 1/50 (2%)
Query 24 IAKHKEDIQEQILKASKNSELPEAREVKEYAMKEIFVP-QTLEEIIEIAN 72
+ + KED+ I+ +N + R + Y +K+ ++P + +E+++ + N
Sbjct 487 LGEQKEDVHYSIISDLQNGDSETRRRLAVYCLKDAYLPLRLMEKLMALVN 536
> Hs19526753
Length=3321
Score = 27.7 bits (60), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 18/40 (45%), Positives = 26/40 (65%), Gaps = 8/40 (20%)
Query 9 DKPVAESSLDDLRSGIAKHKE------DIQEQILKASKNS 42
DKPVA S++ +L++ I++H+E QEQI AS NS
Sbjct 287 DKPVATSNIQELQAQISRHEELAQKIKGYQEQI--ASLNS 324
> Hs5803213
Length=527
Score = 27.7 bits (60), Expect = 5.0, Method: Composition-based stats.
Identities = 12/33 (36%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 60 VPQTLEEIIEIANKDGVVPFTVSAGTPEKKEEK 92
+P+ +EE I+ + K+GVV F++ + EEK
Sbjct 286 LPKEMEEFIQSSGKNGVVVFSLGSMVKNLTEEK 318
> Hs20481728
Length=1498
Score = 27.7 bits (60), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 9/14 (64%), Positives = 14/14 (100%), Gaps = 0/14 (0%)
Query 75 GVVPFTVSAGTPEK 88
GV+PFT+++G+PEK
Sbjct 1374 GVLPFTLNSGSPEK 1387
> 7292158
Length=1240
Score = 27.3 bits (59), Expect = 7.7, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 27/47 (57%), Gaps = 2/47 (4%)
Query 29 EDIQEQILKASKNSELPEAREVKEYAMKEIFVPQTLEEIIEIANKDG 75
+D Q++IL SK S + E+K ++E+F Q L EI + A ++
Sbjct 888 KDAQQEILLPSKASADAKVNEIKSTMLEEVF--QELHEIFQDARENA 932
> YJL090c
Length=764
Score = 27.3 bits (59), Expect = 7.8, Method: Composition-based stats.
Identities = 16/86 (18%), Positives = 35/86 (40%), Gaps = 7/86 (8%)
Query 25 AKHKEDIQEQILKASKNSELPEAREVKEYAMKEIFVPQTLEEIIEIANKDGVVPFTVSAG 84
+ K + I+ S ++P+ + +K ++ + FVP+ ++ +S
Sbjct 655 GRKKRSVSSSIMDVSSERQMPDTKRIKLESLPKNFVPKQIKRTTSWGT-------IMSEN 707
Query 85 TPEKKEEKPVVPVPIPRTRKPTADHV 110
P ++ P IPRT + + V
Sbjct 708 VPTEQPTAISNPEEIPRTEEVSHTQV 733
> Hs18378735
Length=3117
Score = 26.9 bits (58), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 17/64 (26%), Positives = 30/64 (46%), Gaps = 1/64 (1%)
Query 42 SELPEAREVKEYAMKEIFVPQTLEEIIEIANKDGVVPFTVSAGTPEKKEEKPVVPVPIPR 101
S LP+ ++ + A+ + F+P + I++ D T T E + +K PV PR
Sbjct 506 SSLPDNKQEENTALNKDFLPIEIRGILDDLQLDSTA-HTAKQDTVELQNQKSSAPVHAPR 564
Query 102 TRKP 105
+ P
Sbjct 565 SHSP 568
> Hs20543026
Length=621
Score = 26.9 bits (58), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 26/42 (61%), Gaps = 0/42 (0%)
Query 2 TSFTIQLDKPVAESSLDDLRSGIAKHKEDIQEQILKASKNSE 43
TS T ++ K E ++++ I K KE+ + ++ +ASKN+E
Sbjct 400 TSCTKEVGKAALEKQIEEINEQIRKEKEEAEARMRQASKNTE 441
> 7298762
Length=683
Score = 26.9 bits (58), Expect = 9.8, Method: Composition-based stats.
Identities = 13/30 (43%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 21 RSGIAKHKEDIQEQILKASKNSELPEAREV 50
R+GIA + Q +IL A KN E E E+
Sbjct 215 RAGIASEAQKYQAKILSADKNCEYDELIEI 244
> CE09308
Length=1081
Score = 26.9 bits (58), Expect = 10.0, Method: Compositional matrix adjust.
Identities = 11/50 (22%), Positives = 30/50 (60%), Gaps = 1/50 (2%)
Query 24 IAKHKEDIQEQILKASKNSELPEAREVKEYAMKEIFVP-QTLEEIIEIAN 72
+++ KED++ I+ + + R + +Y +K+ ++P + L++++ I N
Sbjct 452 LSEQKEDVEHNIIPDLQRGDEQTRRRLAQYCLKDAYLPLRLLDKLMSIIN 501
> At5g51470
Length=581
Score = 26.9 bits (58), Expect = 10.0, Method: Composition-based stats.
Identities = 22/86 (25%), Positives = 43/86 (50%), Gaps = 8/86 (9%)
Query 3 SFTIQLDKPVAESSLDDLRSGIAKHKEDIQEQILKASKNSELPEAREVKEYAMKEIFVPQ 62
S L + + E L+DL S + + ++++ E+IL + N+E R + + KE+F
Sbjct 2 SLCSDLTEKLDEEILEDLTSNVKQIQDNVLEEILTLNANTEYLR-RFLHGSSSKELF--- 57
Query 63 TLEEIIEIANKDGVVPFT--VSAGTP 86
++ + + + V PF V+ G P
Sbjct 58 --KKNVPVVTYEDVKPFIDRVTNGEP 81
Lambda K H
0.308 0.128 0.343
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1192473466
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40