bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2585_orf4
Length=87
Score E
Sequences producing significant alignments: (Bits) Value
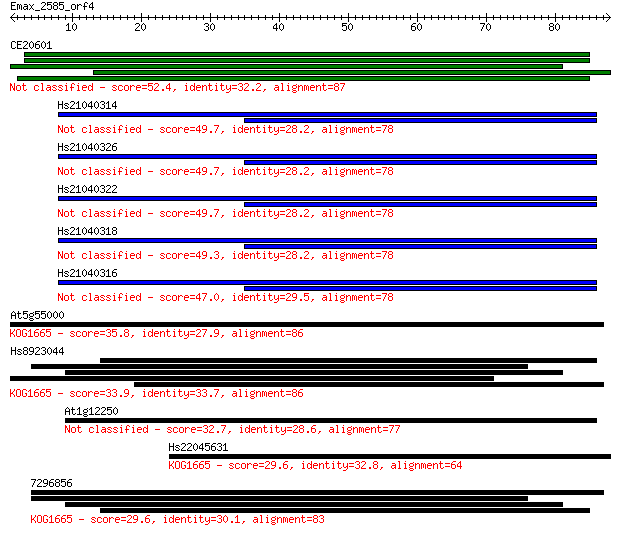
CE20601 52.4 2e-07
Hs21040314 49.7 1e-06
Hs21040326 49.7 1e-06
Hs21040322 49.7 1e-06
Hs21040318 49.3 2e-06
Hs21040316 47.0 8e-06
At5g55000 35.8 0.022
Hs8923044 33.9 0.070
At1g12250 32.7 0.18
Hs22045631 29.6 1.4
7296856 29.6 1.5
> CE20601
Length=726
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 27/82 (32%), Positives = 49/82 (59%), Gaps = 0/82 (0%)
Query 3 VLAGSVLAGSVLAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLAGS 62
+ + VL +VL+ +V + VL+ +V++ V++ +L+ VL VL+ +VL+G
Sbjct 639 IFSPYVLGPNVLSAAVFNAYVFSPYVLSPNVINPYVMSPLILSPFVLCPDVLSPTVLSGV 698
Query 63 VLAGSVLAGSVLAGSVLAGSVL 84
VL+ SVL+ S+ S LA ++L
Sbjct 699 VLSPSVLSPSIFTDSALAANIL 720
Score = 50.8 bits (120), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 28/82 (34%), Positives = 48/82 (58%), Gaps = 0/82 (0%)
Query 3 VLAGSVLAGSVLAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLAGS 62
VL +VL+ +V V + L+ +V+ V+ +L+ VL VL+ +VL+G VL+ S
Sbjct 644 VLGPNVLSAAVFNAYVFSPYVLSPNVINPYVMSPLILSPFVLCPDVLSPTVLSGVVLSPS 703
Query 63 VLAGSVLAGSVLAGSVLAGSVL 84
VL+ S+ S LA ++L+ + L
Sbjct 704 VLSPSIFTDSALAANILSPTFL 725
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 26/80 (32%), Positives = 47/80 (58%), Gaps = 0/80 (0%)
Query 1 GSVLAGSVLAGSVLAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLA 60
+VL+ +V V + VL+ + + V++ +L VL VL+ +VL+G VL+ SVL+
Sbjct 647 PNVLSAAVFNAYVFSPYVLSPNVINPYVMSPLILSPFVLCPDVLSPTVLSGVVLSPSVLS 706
Query 61 GSVLAGSVLAGSVLAGSVLA 80
S+ S LA ++L+ + L+
Sbjct 707 PSIFTDSALAANILSPTFLS 726
Score = 43.1 bits (100), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 24/75 (32%), Positives = 42/75 (56%), Gaps = 0/75 (0%)
Query 13 VLAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGS 72
+ + VL + L+ +V V VL+ +V+ V++ +L+ VL VL+ +VL+G
Sbjct 639 IFSPYVLGPNVLSAAVFNAYVFSPYVLSPNVINPYVMSPLILSPFVLCPDVLSPTVLSGV 698
Query 73 VLAGSVLAGSVLDGS 87
VL+ SVL+ S+ S
Sbjct 699 VLSPSVLSPSIFTDS 713
Score = 32.0 bits (71), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 22/83 (26%), Positives = 44/83 (53%), Gaps = 0/83 (0%)
Query 2 SVLAGSVLAGSVLAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLAG 61
SVL V++ + SVL+ ++ V++ V + + + VL +VL+ +V V +
Sbjct 603 SVLGPLVISPQLFCPSVLSPLLMSLPVISPQVGNPLIFSPYVLGPNVLSAAVFNAYVFSP 662
Query 62 SVLAGSVLAGSVLAGSVLAGSVL 84
VL+ +V+ V++ +L+ VL
Sbjct 663 YVLSPNVINPYVMSPLILSPFVL 685
> Hs21040314
Length=2303
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 42/78 (53%), Gaps = 0/78 (0%)
Query 8 VLAGSVLAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGS 67
+LA + + +LA + + +LA + +D +LA + + +LA S + +LA S +
Sbjct 715 ILASNTMETHILASNTMDSQMLASNTMDSQMLASNTMDSQMLASSTMDSQMLATSSMDSQ 774
Query 68 VLAGSVLAGSVLAGSVLD 85
+LA S + +LA S +D
Sbjct 775 MLATSSMDSQMLATSTMD 792
Score = 33.9 bits (76), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 12/51 (23%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 35 DGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLD 85
+ +LA + + +LA + + +LA + + +LA + + +LA S +D
Sbjct 712 ESHILASNTMETHILASNTMDSQMLASNTMDSQMLASNTMDSQMLASSTMD 762
> Hs21040326
Length=2426
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 42/78 (53%), Gaps = 0/78 (0%)
Query 8 VLAGSVLAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGS 67
+LA + + +LA + + +LA + +D +LA + + +LA S + +LA S +
Sbjct 715 ILASNTMETHILASNTMDSQMLASNTMDSQMLASNTMDSQMLASSTMDSQMLATSSMDSQ 774
Query 68 VLAGSVLAGSVLAGSVLD 85
+LA S + +LA S +D
Sbjct 775 MLATSSMDSQMLATSTMD 792
Score = 33.9 bits (76), Expect = 0.069, Method: Compositional matrix adjust.
Identities = 12/51 (23%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 35 DGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLD 85
+ +LA + + +LA + + +LA + + +LA + + +LA S +D
Sbjct 712 ESHILASNTMETHILASNTMDSQMLASNTMDSQMLASNTMDSQMLASSTMD 762
> Hs21040322
Length=2325
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 42/78 (53%), Gaps = 0/78 (0%)
Query 8 VLAGSVLAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGS 67
+LA + + +LA + + +LA + +D +LA + + +LA S + +LA S +
Sbjct 715 ILASNTMETHILASNTMDSQMLASNTMDSQMLASNTMDSQMLASSTMDSQMLATSSMDSQ 774
Query 68 VLAGSVLAGSVLAGSVLD 85
+LA S + +LA S +D
Sbjct 775 MLATSSMDSQMLATSTMD 792
Score = 33.9 bits (76), Expect = 0.072, Method: Compositional matrix adjust.
Identities = 12/51 (23%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 35 DGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLD 85
+ +LA + + +LA + + +LA + + +LA + + +LA S +D
Sbjct 712 ESHILASNTMETHILASNTMDSQMLASNTMDSQMLASNTMDSQMLASSTMD 762
> Hs21040318
Length=2108
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 22/78 (28%), Positives = 42/78 (53%), Gaps = 0/78 (0%)
Query 8 VLAGSVLAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGS 67
+LA + + +LA + + +LA + +D +LA + + +LA S + +LA S +
Sbjct 715 ILASNTMETHILASNTMDSQMLASNTMDSQMLASNTMDSQMLASSTMDSQMLATSSMDSQ 774
Query 68 VLAGSVLAGSVLAGSVLD 85
+LA S + +LA S +D
Sbjct 775 MLATSSMDSQMLATSTMD 792
Score = 33.9 bits (76), Expect = 0.080, Method: Compositional matrix adjust.
Identities = 12/51 (23%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 35 DGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLD 85
+ +LA + + +LA + + +LA + + +LA + + +LA S +D
Sbjct 712 ESHILASNTMETHILASNTMDSQMLASNTMDSQMLASNTMDSQMLASSTMD 762
> Hs21040316
Length=2386
Score = 47.0 bits (110), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 23/78 (29%), Positives = 42/78 (53%), Gaps = 0/78 (0%)
Query 8 VLAGSVLAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGS 67
+LA + + +LA + + +LA + +D +LA S + +LA S + +LA S +
Sbjct 715 ILASNTMETHILASNTMDSQMLASNTMDSQMLATSTMDSQMLATSSMDSQMLATSSMDSQ 774
Query 68 VLAGSVLAGSVLAGSVLD 85
+LA S + +LA S +D
Sbjct 775 MLATSSMDSQMLATSSMD 792
Score = 33.1 bits (74), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 13/51 (25%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 35 DGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLD 85
+ +LA + + +LA + + +LA + + +LA S + +LA S +D
Sbjct 712 ESHILASNTMETHILASNTMDSQMLASNTMDSQMLATSTMDSQMLATSSMD 762
> At5g55000
Length=298
Score = 35.8 bits (81), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 24/86 (27%), Positives = 41/86 (47%), Gaps = 0/86 (0%)
Query 1 GSVLAGSVLAGSVLAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLA 60
GS+ ++L + L G+ LAG+ L + L + L G G+ L + L + L
Sbjct 180 GSIFHNAILRECEFTSANLRGALLAGTNLQSANLQDACLVGCSFCGADLRTAHLQNADLT 239
Query 61 GSVLAGSVLAGSVLAGSVLAGSVLDG 86
+ L G+ L G+ L G+ L+ + G
Sbjct 240 NANLEGANLEGANLKGAKLSNANFKG 265
> Hs8923044
Length=148
Score = 33.9 bits (76), Expect = 0.070, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 36/72 (50%), Gaps = 0/72 (0%)
Query 14 LAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSV 73
L G+ L G + GS + G L + L + L L G+ LAG+ L L+G L +
Sbjct 64 LEGANLKGVDMEGSQMTGINLRVATLKNAKLKNCNLRGATLAGTDLENCDLSGCDLQEAN 123
Query 74 LAGSVLAGSVLD 85
L GS + G++ +
Sbjct 124 LRGSNVKGAIFE 135
Score = 33.5 bits (75), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 35/72 (48%), Gaps = 0/72 (0%)
Query 9 LAGSVLAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSV 68
L G+ L G + GS + G L + L + L L G+ LAG+ L L+G L +
Sbjct 64 LEGANLKGVDMEGSQMTGINLRVATLKNAKLKNCNLRGATLAGTDLENCDLSGCDLQEAN 123
Query 69 LAGSVLAGSVLA 80
L GS + G++
Sbjct 124 LRGSNVKGAIFE 135
Score = 33.5 bits (75), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 23/72 (31%), Positives = 35/72 (48%), Gaps = 0/72 (0%)
Query 4 LAGSVLAGSVLAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLAGSV 63
L G+ L G + GS + G L + L + L L G+ LAG+ L L+G L +
Sbjct 64 LEGANLKGVDMEGSQMTGINLRVATLKNAKLKNCNLRGATLAGTDLENCDLSGCDLQEAN 123
Query 64 LAGSVLAGSVLA 75
L GS + G++
Sbjct 124 LRGSNVKGAIFE 135
Score = 30.8 bits (68), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 22/70 (31%), Positives = 34/70 (48%), Gaps = 0/70 (0%)
Query 1 GSVLAGSVLAGSVLAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLA 60
G+ L G + GS + G L + L + L L G+ LAG+ L L+G L + L
Sbjct 66 GANLKGVDMEGSQMTGINLRVATLKNAKLKNCNLRGATLAGTDLENCDLSGCDLQEANLR 125
Query 61 GSVLAGSVLA 70
GS + G++
Sbjct 126 GSNVKGAIFE 135
Score = 30.4 bits (67), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 33/68 (48%), Gaps = 0/68 (0%)
Query 19 LAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSV 78
L G+ L G + GS + G L + L + L L G+ LAG+ L L+G L +
Sbjct 64 LEGANLKGVDMEGSQMTGINLRVATLKNAKLKNCNLRGATLAGTDLENCDLSGCDLQEAN 123
Query 79 LAGSVLDG 86
L GS + G
Sbjct 124 LRGSNVKG 131
> At1g12250
Length=280
Score = 32.7 bits (73), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 22/77 (28%), Positives = 40/77 (51%), Gaps = 0/77 (0%)
Query 9 LAGSVLAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSV 68
+ S +GS G+ L +V + G+ L+ +++ VL + L +VL SVL S
Sbjct 136 MRESDFSGSTFNGAYLEKAVAYKANFSGADLSDTLMDRMVLNEANLTNAVLVRSVLTRSD 195
Query 69 LAGSVLAGSVLAGSVLD 85
L G+ + G+ + +V+D
Sbjct 196 LGGAKIEGADFSDAVID 212
> Hs22045631
Length=110
Score = 29.6 bits (65), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 31/64 (48%), Gaps = 0/64 (0%)
Query 24 LAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSV 83
L G+ L G ++GS + G L + L + L L G+ L G+ L L+G L +
Sbjct 26 LEGANLKGVDMEGSQMTGINLRVATLKNAKLKNCSLRGATLTGTDLENCDLSGCDLQEAN 85
Query 84 LDGS 87
L GS
Sbjct 86 LRGS 89
> 7296856
Length=335
Score = 29.6 bits (65), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 38/83 (45%), Gaps = 0/83 (0%)
Query 4 LAGSVLAGSVLAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLAGSV 63
+ G+ L G + L G L G+ L+ S +AG L + L + + L +V
Sbjct 231 MEGANLRGCNFEDPTGVRTNLEGVNLKGACLESSNMAGVNLRVANLKNANMKNCNLRAAV 290
Query 64 LAGSVLAGSVLAGSVLAGSVLDG 86
LAG+ L L+GS L + L G
Sbjct 291 LAGADLEKCNLSGSDLQEANLRG 313
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 35/72 (48%), Gaps = 0/72 (0%)
Query 4 LAGSVLAGSVLAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLAGSV 63
L G L G+ L S +AG L + L + + L +VLAG+ L L+GS L +
Sbjct 251 LEGVNLKGACLESSNMAGVNLRVANLKNANMKNCNLRAAVLAGADLEKCNLSGSDLQEAN 310
Query 64 LAGSVLAGSVLA 75
L G+ L + L
Sbjct 311 LRGANLKDAELT 322
Score = 29.3 bits (64), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 35/72 (48%), Gaps = 0/72 (0%)
Query 9 LAGSVLAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSV 68
L G L G+ L S +AG L + L + + L +VLAG+ L L+GS L +
Sbjct 251 LEGVNLKGACLESSNMAGVNLRVANLKNANMKNCNLRAAVLAGADLEKCNLSGSDLQEAN 310
Query 69 LAGSVLAGSVLA 80
L G+ L + L
Sbjct 311 LRGANLKDAELT 322
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 24/71 (33%), Positives = 35/71 (49%), Gaps = 0/71 (0%)
Query 14 LAGSVLAGSALAGSVLAGSVLDGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSVLAGSV 73
L G L G+ L S +AG L + L + + L +VLAG+ L L+GS L +
Sbjct 251 LEGVNLKGACLESSNMAGVNLRVANLKNANMKNCNLRAAVLAGADLEKCNLSGSDLQEAN 310
Query 74 LAGSVLAGSVL 84
L G+ L + L
Sbjct 311 LRGANLKDAEL 321
Lambda K H
0.310 0.129 0.325
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187582654
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40