bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
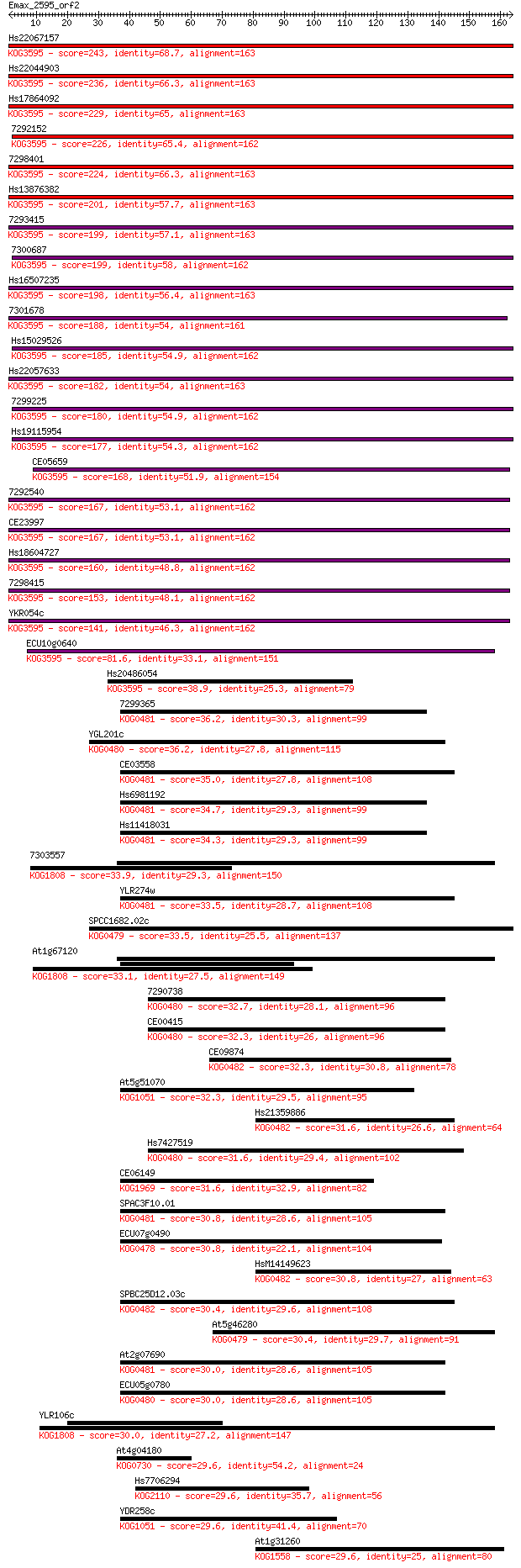
Query= Emax_2595_orf2
Length=163
Score E
Sequences producing significant alignments: (Bits) Value
Hs22067157 243 1e-64
Hs22044903 236 1e-62
Hs17864092 229 1e-60
7292152 226 1e-59
7298401 224 7e-59
Hs13876382 201 6e-52
7293415 199 1e-51
7300687 199 1e-51
Hs16507235 198 4e-51
7301678 188 3e-48
Hs15029526 185 4e-47
Hs22057633 182 4e-46
7299225 180 1e-45
Hs19115954 177 7e-45
CE05659 168 6e-42
7292540 167 6e-42
CE23997 167 1e-41
Hs18604727 160 8e-40
7298415 153 1e-37
YKR054c 141 7e-34
ECU10g0640 81.6 6e-16
Hs20486054 38.9 0.004
7299365 36.2 0.026
YGL201c 36.2 0.031
CE03558 35.0 0.073
Hs6981192 34.7 0.095
Hs11418031 34.3 0.11
7303557 33.9 0.14
YLR274w 33.5 0.17
SPCC1682.02c 33.5 0.21
At1g67120 33.1 0.22
7290738 32.7 0.31
CE00415 32.3 0.37
CE09874 32.3 0.40
At5g51070 32.3 0.47
Hs21359886 31.6 0.72
Hs7427519 31.6 0.73
CE06149 31.6 0.78
SPAC3F10.01 30.8 1.2
ECU07g0490 30.8 1.3
HsM14149623 30.8 1.3
SPBC25D12.03c 30.4 1.5
At5g46280 30.4 1.6
At2g07690 30.0 1.9
ECU05g0780 30.0 2.1
YLR106c 30.0 2.4
At4g04180 29.6 2.5
Hs7706294 29.6 2.6
YDR258c 29.6 2.7
At1g31260 29.6 2.8
> Hs22067157
Length=3672
Score = 243 bits (620), Expect = 1e-64, Method: Composition-based stats.
Identities = 112/163 (68%), Positives = 129/163 (79%), Gaps = 0/163 (0%)
Query 1 FELLRSPDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLV 60
+E L + RLV+TPLTDRCYRTLM AL GGAPEGPAGTGKTETTKDLAKA + C+V
Sbjct 949 YEYLGNSPRLVITPLTDRCYRTLMGALKLNLGGAPEGPAGTGKTETTKDLAKALAKQCVV 1008
Query 61 FNCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITF 120
FNCS+GLD AM KGLA +G W CFDEFNR++++VLS+VA QI +IQQAI RK TF
Sbjct 1009 FNCSDGLDYKAMGKFFKGLAQAGAWACFDEFNRIEVEVLSVVAQQILSIQQAIIRKLKTF 1068
Query 121 VFEDTDLQLNPTCAINITMNPGYAGRSILPDTLKALFRPCTMM 163
+FE T+L LNPTCA+ ITMNPGYAGR+ LPD LKALFR MM
Sbjct 1069 IFEGTELSLNPTCAVFITMNPGYAGRAELPDNLKALFRTVAMM 1111
> Hs22044903
Length=2252
Score = 236 bits (603), Expect = 1e-62, Method: Composition-based stats.
Identities = 108/163 (66%), Positives = 127/163 (77%), Gaps = 0/163 (0%)
Query 1 FELLRSPDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLV 60
+E L + RLV+TPLTDRCYRTL+ A + GGAPEGPAGTGKTETTKDLAKA C+V
Sbjct 838 YEYLGNSPRLVITPLTDRCYRTLIGAFYLNLGGAPEGPAGTGKTETTKDLAKALAVQCVV 897
Query 61 FNCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITF 120
FNCS+GLD AM KGLA+SG W CFDEFNR++L+VLS+VA QI IQ+AI++K + F
Sbjct 898 FNCSDGLDYLAMGKFFKGLASSGAWACFDEFNRIELEVLSVVAQQILCIQRAIQQKLVVF 957
Query 121 VFEDTDLQLNPTCAINITMNPGYAGRSILPDTLKALFRPCTMM 163
VFE T+L+LNP C + ITMNPGYAGRS LPD LK LFR MM
Sbjct 958 VFEGTELKLNPNCFVAITMNPGYAGRSELPDNLKVLFRTVAMM 1000
> Hs17864092
Length=4024
Score = 229 bits (585), Expect = 1e-60, Method: Composition-based stats.
Identities = 106/163 (65%), Positives = 123/163 (75%), Gaps = 0/163 (0%)
Query 1 FELLRSPDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLV 60
+E L + RLV+TPLTDRCYRTL ALH GGAPEGPAGTGKTETTKDLAKA + C+V
Sbjct 1292 YEYLGNSPRLVITPLTDRCYRTLFGALHLHLGGAPEGPAGTGKTETTKDLAKAVAKQCVV 1351
Query 61 FNCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITF 120
FNCS+GLD A+ KGL + G W CFDEFNR+ L+VLS+VA QI IQ+ I A
Sbjct 1352 FNCSDGLDYLALGKFFKGLLSCGAWACFDEFNRIDLEVLSVVAQQILTIQRGINAGADIL 1411
Query 121 VFEDTDLQLNPTCAINITMNPGYAGRSILPDTLKALFRPCTMM 163
+FE T+L+L+PTCA+ ITMNPGYAGRS LPD LKALFR MM
Sbjct 1412 MFEGTELKLDPTCAVFITMNPGYAGRSELPDNLKALFRTVAMM 1454
> 7292152
Length=3868
Score = 226 bits (577), Expect = 1e-59, Method: Composition-based stats.
Identities = 106/162 (65%), Positives = 119/162 (73%), Gaps = 0/162 (0%)
Query 2 ELLRSPDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLVF 61
E L + DRLV+TPLTDRCYRTL+ A GAPEGPAGTGKTETTKDLAKA C VF
Sbjct 1242 EYLGNSDRLVITPLTDRCYRTLVGAYQLHLNGAPEGPAGTGKTETTKDLAKALAVQCKVF 1301
Query 62 NCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFV 121
NCS+GLD AM KGLA+ G W CFDEFNR++L+VLS+VA QI I QA+R A F+
Sbjct 1302 NCSDGLDYKAMGKFFKGLASCGAWACFDEFNRIELEVLSVVAQQILLIIQAVRSNATKFM 1361
Query 122 FEDTDLQLNPTCAINITMNPGYAGRSILPDTLKALFRPCTMM 163
FE T+L LNP C + ITMNPGYAGRS LPD LK LFR MM
Sbjct 1362 FEGTELTLNPACYVCITMNPGYAGRSELPDNLKVLFRSVAMM 1403
> 7298401
Length=4010
Score = 224 bits (570), Expect = 7e-59, Method: Compositional matrix adjust.
Identities = 108/163 (66%), Positives = 123/163 (75%), Gaps = 0/163 (0%)
Query 1 FELLRSPDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLV 60
+E L + RLVVTPLTDRCYRTL AL+ GGAPEGPAGTGKTETTKDLAKA + C+V
Sbjct 1262 YEYLGNTPRLVVTPLTDRCYRTLFAALNLHLGGAPEGPAGTGKTETTKDLAKAVAKQCVV 1321
Query 61 FNCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITF 120
FNCS+GLD A+ KGLA+ G W CFDEFNR+ L+VLS+VA QI IQ+ I + T
Sbjct 1322 FNCSDGLDYLALGKFFKGLASCGAWSCFDEFNRIDLEVLSVVAQQILTIQRGINSGSPTL 1381
Query 121 VFEDTDLQLNPTCAINITMNPGYAGRSILPDTLKALFRPCTMM 163
VFE T L L+PTCA+ ITMNPGYAGRS LPD LKALFR MM
Sbjct 1382 VFEGTTLTLDPTCAVFITMNPGYAGRSELPDNLKALFRSVAMM 1424
> Hs13876382
Length=4486
Score = 201 bits (510), Expect = 6e-52, Method: Compositional matrix adjust.
Identities = 94/163 (57%), Positives = 117/163 (71%), Gaps = 0/163 (0%)
Query 1 FELLRSPDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLV 60
+E L + RLV+TPLTDRCY TL +LH GAP GPAGTGKTETTKDL +A G V
Sbjct 1834 YEYLGNTPRLVITPLTDRCYITLTQSLHLTMSGAPAGPAGTGKTETTKDLGRALGILVYV 1893
Query 61 FNCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITF 120
FNCSE +D + + KGLA +G W CFDEFNR+ ++VLS+VA+Q+ +IQ AIR K F
Sbjct 1894 FNCSEQMDYKSCGNIYKGLAQTGAWGCFDEFNRISVEVLSVVAVQVKSIQDAIRDKKQWF 1953
Query 121 VFEDTDLQLNPTCAINITMNPGYAGRSILPDTLKALFRPCTMM 163
F ++ LNP+ I ITMNPGYAGR+ LP+ LK+LFRPC M+
Sbjct 1954 SFLGEEISLNPSVGIFITMNPGYAGRTELPENLKSLFRPCAMV 1996
> 7293415
Length=4081
Score = 199 bits (507), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 93/163 (57%), Positives = 116/163 (71%), Gaps = 0/163 (0%)
Query 1 FELLRSPDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLV 60
+E L + LV+TPLTDRCY LM A GGAP GPAGTGKTETTKDLAKA + C+V
Sbjct 1375 YEYLGAGGVLVLTPLTDRCYLCLMGAFQMDLGGAPAGPAGTGKTETTKDLAKALAKQCVV 1434
Query 61 FNCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITF 120
FNCS+GLD M GLA G WCCFDEFNR+ ++VLS++A Q+ I+ A + F
Sbjct 1435 FNCSDGLDYKMMGRFFSGLAQCGAWCCFDEFNRIDIEVLSVIAQQLITIRTAKAMRVKRF 1494
Query 121 VFEDTDLQLNPTCAINITMNPGYAGRSILPDTLKALFRPCTMM 163
+FE ++++N +C + ITMNPGYAGR+ LPD LKALFRP +MM
Sbjct 1495 IFEGREIKINRSCCVFITMNPGYAGRTELPDNLKALFRPISMM 1537
> 7300687
Length=4472
Score = 199 bits (507), Expect = 1e-51, Method: Composition-based stats.
Identities = 94/162 (58%), Positives = 114/162 (70%), Gaps = 0/162 (0%)
Query 2 ELLRSPDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLVF 61
E L + RLV+TPLTDRCY TL +LH GGAP GPAGTGKTETTKDL +A G VF
Sbjct 1832 EYLGNTPRLVITPLTDRCYITLTQSLHLVMGGAPAGPAGTGKTETTKDLGRAIGISVYVF 1891
Query 62 NCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFV 121
NCSE +D + + KGLA +G W CFDEFNR+ ++VLS+VA+Q+ ++Q AIR K F
Sbjct 1892 NCSEQMDYQSCGNIYKGLAQTGAWGCFDEFNRITVEVLSVVAVQVKSVQDAIRDKKDKFN 1951
Query 122 FEDTDLQLNPTCAINITMNPGYAGRSILPDTLKALFRPCTMM 163
F + PT I ITMNPGYAGR+ LP+ LKALFRPC M+
Sbjct 1952 FMGEMISCVPTVGIFITMNPGYAGRTELPENLKALFRPCAMV 1993
> Hs16507235
Length=4523
Score = 198 bits (503), Expect = 4e-51, Method: Compositional matrix adjust.
Identities = 92/163 (56%), Positives = 114/163 (69%), Gaps = 0/163 (0%)
Query 1 FELLRSPDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLV 60
+E L + RLV+TPLTDRCY TL +LH GAP GPAGTGKTETTKDL +A G V
Sbjct 1864 YEYLGNSPRLVITPLTDRCYITLTQSLHLTMSGAPAGPAGTGKTETTKDLGRALGMMVYV 1923
Query 61 FNCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITF 120
FNCSE +D ++ + KGL +G W CFDEFNR+ ++VLS+VA+Q+ I AIR + F
Sbjct 1924 FNCSEQMDYKSIGNIYKGLVQTGAWGCFDEFNRISVEVLSVVAVQVKMIHDAIRNRKKRF 1983
Query 121 VFEDTDLQLNPTCAINITMNPGYAGRSILPDTLKALFRPCTMM 163
VF + L P+ I ITMNPGYAGR+ LP+ LKALFRPC M+
Sbjct 1984 VFLGEAITLKPSVGIFITMNPGYAGRTELPENLKALFRPCAMV 2026
> 7301678
Length=4820
Score = 188 bits (478), Expect = 3e-48, Method: Compositional matrix adjust.
Identities = 87/161 (54%), Positives = 112/161 (69%), Gaps = 0/161 (0%)
Query 1 FELLRSPDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLV 60
+E + RLV+TPLTDR Y T+ AL GGAP GPAGTGKTET KDLAKA G C+V
Sbjct 2391 YEYMGLNGRLVITPLTDRIYLTITQALLMNLGGAPAGPAGTGKTETVKDLAKAMGLLCVV 2450
Query 61 FNCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITF 120
NC EG+D A+ +L GL G W CFDEFNR+ + VLS+++ Q+ I+ + RK F
Sbjct 2451 TNCGEGMDYRAVGTILSGLVQCGAWGCFDEFNRIDISVLSVISTQLQTIRNGLIRKLDRF 2510
Query 121 VFEDTDLQLNPTCAINITMNPGYAGRSILPDTLKALFRPCT 161
VFE ++ L+P C + +TMNPGYAGR+ LP+++KALFRP T
Sbjct 2511 VFEGVEIHLDPKCGVFVTMNPGYAGRTELPESVKALFRPVT 2551
> Hs15029526
Length=4490
Score = 185 bits (469), Expect = 4e-47, Method: Composition-based stats.
Identities = 89/163 (54%), Positives = 108/163 (66%), Gaps = 1/163 (0%)
Query 2 ELLRSPDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLVF 61
E L DRLV+TPLTDRCY TL AL GGAP GPAGTGKTETTKD+ + G+ +VF
Sbjct 1811 EFLGCTDRLVITPLTDRCYITLAQALGMNMGGAPAGPAGTGKTETTKDMGRCLGKYVVVF 1870
Query 62 NCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFV 121
NCS+ +D + + KGLA SG W CFDEFNR++L VLS+ A QI + A + + F+
Sbjct 1871 NCSDQMDFRGLGRIFKGLAQSGSWGCFDEFNRIELPVLSVAAQQIYIVLTARKERKKQFI 1930
Query 122 FEDTD-LQLNPTCAINITMNPGYAGRSILPDTLKALFRPCTMM 163
F D D + LNP I +TMNPGYAGR LP+ LK FR MM
Sbjct 1931 FSDGDCVDLNPEFGIFLTMNPGYAGRQELPENLKIQFRTVAMM 1973
> Hs22057633
Length=1770
Score = 182 bits (461), Expect = 4e-46, Method: Compositional matrix adjust.
Identities = 88/163 (53%), Positives = 111/163 (68%), Gaps = 0/163 (0%)
Query 1 FELLRSPDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLV 60
+E + RLV+TPLTDR Y TL AL GGAP GPAGTGKTETTKDLAKA G C+V
Sbjct 279 YEYMGLNGRLVITPLTDRIYLTLTQALSMYLGGAPAGPAGTGKTETTKDLAKALGLLCVV 338
Query 61 FNCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITF 120
NC EG+D A+ + GLA G W CFDEFNR+ VLS+++ QI I+ A+ + TF
Sbjct 339 TNCGEGMDYRAVGKIFSGLAQCGAWGCFDEFNRIDASVLSVISSQIQTIRNALIHQLTTF 398
Query 121 VFEDTDLQLNPTCAINITMNPGYAGRSILPDTLKALFRPCTMM 163
FE ++ L+ I ITMNPGYAGR+ LP+++KALFRP ++
Sbjct 399 QFEGQEISLDSRMGIFITMNPGYAGRTELPESVKALFRPVVVI 441
> 7299225
Length=3508
Score = 180 bits (457), Expect = 1e-45, Method: Compositional matrix adjust.
Identities = 89/163 (54%), Positives = 109/163 (66%), Gaps = 1/163 (0%)
Query 2 ELLRSPDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLVF 61
E L DRLV+TPLTDRCY TL AL GGAP GPAGTGKTET KD+ K + +VF
Sbjct 1926 EYLGCTDRLVITPLTDRCYITLAQALTLSMGGAPCGPAGTGKTETVKDMGKTLAKYVVVF 1985
Query 62 NCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFV 121
NCS+ +D + + KGLA SG W CFDEFNR++L VLS+ A Q++ + A + K TF+
Sbjct 1986 NCSDQMDYRGLGRIYKGLAQSGSWGCFDEFNRIELPVLSVAAQQVAVVLTAKKEKRKTFL 2045
Query 122 FEDTD-LQLNPTCAINITMNPGYAGRSILPDTLKALFRPCTMM 163
F D D +++NP I ITMNPGYAGR LP+ LK FR MM
Sbjct 2046 FTDGDTIEMNPEFGIFITMNPGYAGRKELPENLKIQFRTVAMM 2088
> Hs19115954
Length=4624
Score = 177 bits (450), Expect = 7e-45, Method: Compositional matrix adjust.
Identities = 88/163 (53%), Positives = 107/163 (65%), Gaps = 1/163 (0%)
Query 2 ELLRSPDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLVF 61
E L DRLV+TPLTDRCY TL AL GGAP GPAGTGKTETTKD+ + G+ +VF
Sbjct 1945 EFLGCTDRLVITPLTDRCYITLAQALGMSMGGAPAGPAGTGKTETTKDMGRCLGKYVVVF 2004
Query 62 NCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFV 121
NCS+ +D + + KGLA SG W CFDEFNR+ L VLS+ A QIS I + +F+
Sbjct 2005 NCSDQMDFRGLGRIFKGLAQSGSWGCFDEFNRIDLPVLSVAAQQISIILTCKKEHKKSFI 2064
Query 122 FEDTD-LQLNPTCAINITMNPGYAGRSILPDTLKALFRPCTMM 163
F D D + +NP + +TMNPGYAGR LP+ LK FR MM
Sbjct 2065 FTDGDNVTMNPEFGLFLTMNPGYAGRQELPENLKINFRSVAMM 2107
> CE05659
Length=4131
Score = 168 bits (425), Expect = 6e-42, Method: Composition-based stats.
Identities = 80/157 (50%), Positives = 100/157 (63%), Gaps = 3/157 (1%)
Query 9 RLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLVFNCSEGLD 68
+LV TPLTD+CY TL A++ GG P GPAGTGKTE+ K LA G LVFNC EG+D
Sbjct 1609 KLVHTPLTDKCYLTLTQAMYMGLGGNPYGPAGTGKTESVKALAALMGRQVLVFNCDEGID 1668
Query 69 AAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFVFEDTDLQ 128
+M + G+ G W CFDEFNRL VLS V++QI IQ AI+ +A + F ++Q
Sbjct 1669 VTSMGRIFTGIVECGAWGCFDEFNRLDSTVLSAVSMQIQTIQGAIKSRAGSCTFGGKNVQ 1728
Query 129 LNPTCAINITMNP---GYAGRSILPDTLKALFRPCTM 162
+NP AI +T+NP GY GR +PD LK LFR M
Sbjct 1729 VNPNSAIFVTLNPAGKGYGGRQKMPDNLKQLFRAVVM 1765
> 7292540
Length=4680
Score = 167 bits (424), Expect = 6e-42, Method: Composition-based stats.
Identities = 86/168 (51%), Positives = 104/168 (61%), Gaps = 6/168 (3%)
Query 1 FELLRSPDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLV 60
FE L DRLV TPLTDRCY T+ AL + GG+P GPAGTGKTE+ K L G LV
Sbjct 1859 FEYLGVQDRLVQTPLTDRCYLTMTQALESRLGGSPFGPAGTGKTESVKALGNQLGRFVLV 1918
Query 61 FNCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIR------ 114
FNC E D AM + GL G W CFDEFNRL+ +LS + QI IQ+A++
Sbjct 1919 FNCDETFDFQAMGRIFVGLCQVGAWGCFDEFNRLEERMLSACSQQIQTIQEALKYEMDSN 1978
Query 115 RKAITFVFEDTDLQLNPTCAINITMNPGYAGRSILPDTLKALFRPCTM 162
+++IT ++++P AI ITMNPGYAGRS LPD LK LFR M
Sbjct 1979 KESITVELVGKQVRVSPDMAIFITMNPGYAGRSNLPDNLKKLFRSLAM 2026
> CE23997
Length=4568
Score = 167 bits (422), Expect = 1e-41, Method: Composition-based stats.
Identities = 86/163 (52%), Positives = 101/163 (61%), Gaps = 1/163 (0%)
Query 1 FELLRSPDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLV 60
FE L +RLV TPLTDRCY T+ ALH + GG+P GPAGTGKTE+ K L G LV
Sbjct 1829 FEYLGIQERLVRTPLTDRCYLTMTQALHSRLGGSPFGPAGTGKTESVKALGHQLGRFVLV 1888
Query 61 FNCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKA-IT 119
FNC E D AM +L GL G W CFDEFNRL+ +LS V+ QI IQ+A+R ++
Sbjct 1889 FNCDETFDFQAMGRILVGLCQVGAWGCFDEFNRLEERMLSAVSQQIQTIQEAVRAGGDMS 1948
Query 120 FVFEDTDLQLNPTCAINITMNPGYAGRSILPDTLKALFRPCTM 162
L +N I ITMNPGY+GRS LPD LK LFR M
Sbjct 1949 VDLVGKRLNVNSNIGIFITMNPGYSGRSNLPDNLKQLFRSLAM 1991
> Hs18604727
Length=2504
Score = 160 bits (406), Expect = 8e-40, Method: Composition-based stats.
Identities = 79/165 (47%), Positives = 97/165 (58%), Gaps = 3/165 (1%)
Query 1 FELLRSPDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLV 60
+E + +LV TPLTD+CY TL A+ GG P GPAGTGKTE+ K L G LV
Sbjct 1559 YEYQGNASKLVYTPLTDKCYLTLTQAMKMGLGGNPYGPAGTGKTESVKALGGLLGRQVLV 1618
Query 61 FNCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITF 120
FNC EG+D +M + GL G W CFDEFNRL+ VLS V++QI IQ A++
Sbjct 1619 FNCDEGIDVKSMGRIFVGLVKCGAWGCFDEFNRLEESVLSAVSMQIQTIQDALKNHRTVC 1678
Query 121 VFEDTDLQLNPTCAINITMNP---GYAGRSILPDTLKALFRPCTM 162
++++N I ITMNP GY GR LPD LK LFRP M
Sbjct 1679 ELLGKEVEVNSNSGIFITMNPAGKGYGGRQKLPDNLKQLFRPVAM 1723
> 7298415
Length=2055
Score = 153 bits (387), Expect = 1e-37, Method: Composition-based stats.
Identities = 78/166 (46%), Positives = 98/166 (59%), Gaps = 4/166 (2%)
Query 1 FELLRSPDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLV 60
+E L +LV T LT RCY L A+H GG P GPAGTGKTE K L G LV
Sbjct 1582 YEFLGQASKLVHTRLTHRCYLILTQAMHMGLGGNPFGPAGTGKTECVKALGAMLGRLVLV 1641
Query 61 FNCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITF 120
FNC E +D +M+ +L GLA G W CFDEFNRLQ LS +++ I IQ A++ +A +
Sbjct 1642 FNCDENVDTESMSLILTGLARCGAWGCFDEFNRLQEATLSSISMLIQPIQSALKERANSV 1701
Query 121 -VFEDTDLQLNPTCAINITMNPG---YAGRSILPDTLKALFRPCTM 162
+ E +QLN C I +T+NP Y GR LP ++ALFRP M
Sbjct 1702 QIGERQQIQLNQHCGIFVTLNPAGAEYGGRQKLPGNIQALFRPIVM 1747
> YKR054c
Length=4092
Score = 141 bits (355), Expect = 7e-34, Method: Composition-based stats.
Identities = 75/164 (45%), Positives = 98/164 (59%), Gaps = 4/164 (2%)
Query 1 FELLRSPDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLV 60
FE + P+RL+ TPL + TL +LH +YGG GPAGTGKTET K + G +V
Sbjct 1760 FEYIGIPERLIYTPLLLIGFATLTDSLHQKYGGCFFGPAGTGKTETVKAFGQNLGRVVVV 1819
Query 61 FNCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIR--RKAI 118
FNC + D ++ LL G+ G W CFDEFNRL VLS V+ I IQ ++ + I
Sbjct 1820 FNCDDSFDYQVLSRLLVGITQIGAWGCFDEFNRLDEKVLSAVSANIQQIQNGLQVGKSHI 1879
Query 119 TFVFEDTDLQLNPTCAINITMNPGYAGRSILPDTLKALFRPCTM 162
T + E+T L+P A+ IT+NPGY GRS LP+ LK FR +M
Sbjct 1880 TLLEEET--PLSPHTAVFITLNPGYNGRSELPENLKKSFREFSM 1921
> ECU10g0640
Length=3151
Score = 81.6 bits (200), Expect = 6e-16, Method: Composition-based stats.
Identities = 50/151 (33%), Positives = 73/151 (48%), Gaps = 20/151 (13%)
Query 7 PDRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLVFNCSEG 66
P +V TPL R ++ +L G G +GTGKTE+ K + G+P VF C+E
Sbjct 1584 PTDIVFTPLVCRVLSSIAVSLKSLCGAILYGRSGTGKTESVKYYCRLIGKPVFVFCCNED 1643
Query 67 LDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFVFEDTD 126
+ A + +++G G + CFDEFNRL + +S A + + K T F
Sbjct 1644 CELATLRNVIEGAVLMGSYLCFDEFNRLSEETMS------GATELILSSKDKTKFF---- 1693
Query 127 LQLNPTCAINITMNPGYAGRSILPDTLKALF 157
+TMN GY GR LP +L+A+F
Sbjct 1694 ----------LTMNIGYKGRYELPRSLRAIF 1714
> Hs20486054
Length=972
Score = 38.9 bits (89), Expect = 0.004, Method: Composition-based stats.
Identities = 20/79 (25%), Positives = 36/79 (45%), Gaps = 0/79 (0%)
Query 33 GAPEGPAGTGKTETTKDLAKAAGEPCLVFNCSEGLDAAAMAALLKGLAASGGWCCFDEFN 92
G GP G GK LA+A G ++ CS ++A ++ L G G W ++ +
Sbjct 341 GTVLGPNGVGKRAIVNSLAQALGRQLVMLPCSPQIEAQCLSNYLNGALQGGAWLLLEKVH 400
Query 93 RLQLDVLSIVALQISAIQQ 111
+L +LS + ++ +
Sbjct 401 QLPPGLLSALGQRLGELHH 419
> 7299365
Length=733
Score = 36.2 bits (82), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 30/113 (26%), Positives = 50/113 (44%), Gaps = 23/113 (20%)
Query 37 GPAGTGKTETTKDLAKAAGEPCLVFNCSEGLDAAAMAA-----------LLKGLA---AS 82
G GT K++ K + K A P V+ +G AA + A +++G A A
Sbjct 378 GDPGTAKSQLLKFVEKVA--PIAVYTSGKGSSAAGLTASVMKDPQTRNFVMEGGAMVLAD 435
Query 83 GGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFVFEDTDLQLNPTCAI 135
GG C DEF++++ D AI +A+ ++ I+ LN C++
Sbjct 436 GGVVCIDEFDKMRED-------DRVAIHEAMEQQTISIAKAGITTTLNSRCSV 481
> YGL201c
Length=1017
Score = 36.2 bits (82), Expect = 0.031, Method: Composition-based stats.
Identities = 32/115 (27%), Positives = 48/115 (41%), Gaps = 15/115 (13%)
Query 27 LHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLVFNCSEGLDAAAMAALLKGLAASGGWC 86
L + G AP +GK + AAG V EG D A L + A G C
Sbjct 585 LKYVVGFAPRSVYTSGKASS------AAGLTAAVVRDEEGGDYTIEAGAL--MLADNGIC 636
Query 87 CFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFVFEDTDLQLNPTCAINITMNP 141
C DEF+++ ++ Q+ AI +A+ ++ I+ LN +I NP
Sbjct 637 CIDEFDKMD------ISDQV-AIHEAMEQQTISIAKAGIHATLNARTSILAAANP 684
> CE03558
Length=759
Score = 35.0 bits (79), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 30/122 (24%), Positives = 52/122 (42%), Gaps = 23/122 (18%)
Query 37 GPAGTGKTETTKDLAKAAGEPCLVFNCSEGLDAAAMAA-----------LLKGLA---AS 82
G GT K++ K + + + P V+ +G AA + A +++G A A
Sbjct 380 GDPGTAKSQLLKFVEQVS--PIGVYTSGKGSSAAGLTASVIRDPQSRSFIMEGGAMVLAD 437
Query 83 GGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFVFEDTDLQLNPTCAINITMNPG 142
GG C DEF++++ D AI +A+ ++ I+ LN C++ N
Sbjct 438 GGVVCIDEFDKMRED-------DRVAIHEAMEQQTISIAKAGITTTLNSRCSVLAAANSV 490
Query 143 YA 144
Y
Sbjct 491 YG 492
> Hs6981192
Length=733
Score = 34.7 bits (78), Expect = 0.095, Method: Compositional matrix adjust.
Identities = 29/113 (25%), Positives = 50/113 (44%), Gaps = 23/113 (20%)
Query 37 GPAGTGKTETTKDLAKAAGEPCLVFNCSEGLDAAAMAA-----------LLKGLA---AS 82
G GT K++ K + K + P V+ +G AA + A +++G A A
Sbjct 381 GDPGTAKSQLLKFVEKCS--PIGVYTSGKGSSAAGLTASVMRDPSSRNFIMEGGAMVLAD 438
Query 83 GGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFVFEDTDLQLNPTCAI 135
GG C DEF++++ D AI +A+ ++ I+ LN C++
Sbjct 439 GGVVCIDEFDKMRED-------DRVAIHEAMEQQTISIAKAGITTTLNSRCSV 484
> Hs11418031
Length=734
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 29/113 (25%), Positives = 50/113 (44%), Gaps = 23/113 (20%)
Query 37 GPAGTGKTETTKDLAKAAGEPCLVFNCSEGLDAAAMAA-----------LLKGLA---AS 82
G GT K++ K + K + P V+ +G AA + A +++G A A
Sbjct 381 GDPGTAKSQLLKFVEKCS--PIGVYTSGKGSSAAGLTASVMRDPSSRNFIMEGGAMVLAD 438
Query 83 GGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFVFEDTDLQLNPTCAI 135
GG C DEF++++ D AI +A+ ++ I+ LN C++
Sbjct 439 GGVVCIDEFDKMRED-------DRVAIHEAMEQQTISIAKAGITTTLNSRCSV 484
> 7303557
Length=4865
Score = 33.9 bits (76), Expect = 0.14, Method: Composition-based stats.
Identities = 36/140 (25%), Positives = 50/140 (35%), Gaps = 25/140 (17%)
Query 36 EGPAGTGKTETTKDLAKAAGEPCLVFNCSEGLD----AAAMAALLKG-----------LA 80
+GP GKT +A+ +G CL N E D AA L G
Sbjct 1115 QGPTSAGKTSLIDYVARRSGNRCLRINNHEHTDLQEYIGTYAADLDGKLTFREGVLVQAM 1174
Query 81 ASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFVFE-DTDLQLNPTCAINITM 139
G W DE N D+L A+ + + ++ E T ++ +P + T
Sbjct 1175 RHGFWIILDELNLASTDIL-------EALNRVLDDNRELYIAETQTLVKAHPNFMLFATQ 1227
Query 140 NPG--YAGRSILPDTLKALF 157
NP Y GR L K F
Sbjct 1228 NPPGLYGGRKTLSRAFKNRF 1247
Score = 33.1 bits (74), Expect = 0.23, Method: Composition-based stats.
Identities = 21/65 (32%), Positives = 28/65 (43%), Gaps = 0/65 (0%)
Query 8 DRLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLVFNCSEGL 67
D L P T + L++AL Q EGP G GKT + + A G + N E
Sbjct 1735 DFLFDAPTTKQNLFRLLSALSLQKPVLLEGPPGVGKTSIVESIGSAIGYQIVRINLCEHT 1794
Query 68 DAAAM 72
D A +
Sbjct 1795 DLADL 1799
> YLR274w
Length=775
Score = 33.5 bits (75), Expect = 0.17, Method: Composition-based stats.
Identities = 31/122 (25%), Positives = 51/122 (41%), Gaps = 23/122 (18%)
Query 37 GPAGTGKTETTKDLAKAAGEPCLVFNCSEGLDAAAMAA-----------LLKGLA---AS 82
G GT K++ K + K + P V+ +G AA + A L+G A A
Sbjct 416 GDPGTAKSQLLKFVEKVS--PIAVYTSGKGSSAAGLTASVQRDPMTREFYLEGGAMVLAD 473
Query 83 GGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFVFEDTDLQLNPTCAINITMNPG 142
GG C DEF++++ + AI +A+ ++ I+ LN ++ NP
Sbjct 474 GGVVCIDEFDKMRDE-------DRVAIHEAMEQQTISIAKAGITTVLNSRTSVLAAANPI 526
Query 143 YA 144
Y
Sbjct 527 YG 528
> SPCC1682.02c
Length=879
Score = 33.5 bits (75), Expect = 0.21, Method: Composition-based stats.
Identities = 35/143 (24%), Positives = 55/143 (38%), Gaps = 27/143 (18%)
Query 27 LHFQYGGAPEGPAGTGKTETTKDLA------KAAGEPCLVFNCSEGLDAAAMAALLKGLA 80
L F AP A TG+ + L K GE L+A AM +
Sbjct 366 LRFVLNTAPLAIATTGRGSSGVGLTAAVTTDKETGE--------RRLEAGAM------VL 411
Query 81 ASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFVFEDTDLQLNPTCAINITMN 140
A G C DEF+++ D+ + AI + + ++ +T LN C++ N
Sbjct 412 ADRGVVCIDEFDKMS-DIDRV------AIHEVMEQQTVTIAKAGIHTSLNARCSVIAAAN 464
Query 141 PGYAGRSILPDTLKALFRPCTMM 163
P Y I D + + P +M+
Sbjct 465 PIYGQYDIRKDPHQNIALPDSML 487
> At1g67120
Length=5138
Score = 33.1 bits (74), Expect = 0.22, Method: Composition-based stats.
Identities = 37/141 (26%), Positives = 52/141 (36%), Gaps = 27/141 (19%)
Query 36 EGPAGTGKTETTKDLAKAAGEPCLVFNCSEGLDAAAM----------------AALLKGL 79
+GP +GKT K LA +G + N E D AL+K +
Sbjct 965 QGPTSSGKTSLVKYLAAISGNKFVRINNHEQTDIQEYLGSYMTDSSGKLVFHEGALVKAV 1024
Query 80 AASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFVFEDTD-LQLNPTCAINIT 138
G W DE N DVL A+ + + FV E ++ + +P + T
Sbjct 1025 RG-GHWIVLDELNLAPSDVL-------EALNRLLDDNRELFVPELSETISAHPNFMLFAT 1076
Query 139 MNPG--YAGRSILPDTLKALF 157
NP Y GR IL + F
Sbjct 1077 QNPPTLYGGRKILSRAFRNRF 1097
Score = 29.3 bits (64), Expect = 3.1, Method: Composition-based stats.
Identities = 15/56 (26%), Positives = 25/56 (44%), Gaps = 12/56 (21%)
Query 37 GPAGTGKTETTKDLAKAAGEPCLVFNCSEGLDAAAMAALLKGLAASGGWCCFDEFN 92
GP+ +GKT + LA+ G P N S D++ + CF+++N
Sbjct 1883 GPSSSGKTSVIRILAQLTGYPLNELNLSSATDSSDLLG------------CFEQYN 1926
Score = 28.9 bits (63), Expect = 4.3, Method: Composition-based stats.
Identities = 24/93 (25%), Positives = 38/93 (40%), Gaps = 5/93 (5%)
Query 9 RLVVTPLTDRCYRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLVFNCSEGLD 68
R V T + R + ++ + G GTGKT ++LA G+ V N S+ D
Sbjct 533 RFVETRTSTRLLEKIARSVEYNEPVLLVGETGTGKTTLVQNLAHWIGQKLTVLNLSQQSD 592
Query 69 AAAMAALLKGLAASGGWCC---FDEFNRLQLDV 98
+ K + C ++EFN L D+
Sbjct 593 IVDLLGGFKPIDPK--LMCTMVYNEFNELARDL 623
> 7290738
Length=817
Score = 32.7 bits (73), Expect = 0.31, Method: Composition-based stats.
Identities = 27/97 (27%), Positives = 40/97 (41%), Gaps = 11/97 (11%)
Query 46 TTKDLAKAAGEPCLVFNCSEGLDAAAMAALLKGLAASGGWCCFDEFNRL-QLDVLSIVAL 104
T+ + AAG V E D A L + A G CC DEF+++ Q D +
Sbjct 411 TSGKASSAAGLTAAVVRDEESFDFVIEAGAL--MLADNGICCIDEFDKMDQRDQV----- 463
Query 105 QISAIQQAIRRKAITFVFEDTDLQLNPTCAINITMNP 141
AI +A+ ++ I+ LN +I NP
Sbjct 464 ---AIHEAMEQQTISIARAGVRATLNARTSILAAANP 497
> CE00415
Length=810
Score = 32.3 bits (72), Expect = 0.37, Method: Composition-based stats.
Identities = 25/96 (26%), Positives = 38/96 (39%), Gaps = 9/96 (9%)
Query 46 TTKDLAKAAGEPCLVFNCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQ 105
T+ + AAG V E + A L + A G CC DEF+++ L
Sbjct 420 TSGKASSAAGLTAAVVKDEESFEFVIEAGAL--MLADNGVCCIDEFDKMDLK-------D 470
Query 106 ISAIQQAIRRKAITFVFEDTDLQLNPTCAINITMNP 141
AI +A+ ++ I+ LN +I NP
Sbjct 471 QVAIHEAMEQQTISITKAGVKATLNARASILAAANP 506
> CE09874
Length=730
Score = 32.3 bits (72), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 24/87 (27%), Positives = 36/87 (41%), Gaps = 16/87 (18%)
Query 66 GLDAAAMAALLKG---------LAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRK 116
GL AA M + G + A GG CC DEF+++ + +AI + + ++
Sbjct 422 GLTAAVMKDPVTGEMSLEGGALVLADGGICCIDEFDKM-------MDHDRTAIHEVMEQQ 474
Query 117 AITFVFEDTDLQLNPTCAINITMNPGY 143
I+ LN AI NP Y
Sbjct 475 TISIAKAGIMTTLNARTAIIAAANPAY 501
> At5g51070
Length=945
Score = 32.3 bits (72), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 28/98 (28%), Positives = 42/98 (42%), Gaps = 21/98 (21%)
Query 37 GPAGTGKTETTKDLAK---AAGEPCLVFNCSEGLDAAAMAALLKGLAASGGWCCFDEFNR 93
GP G GKTE TK LA + E L + SE ++ ++ L + + G+ F+E
Sbjct 664 GPTGVGKTELTKALAANYFGSEESMLRLDMSEYMERHTVSKL---IGSPPGYVGFEEGGM 720
Query 94 LQLDVLSIVALQISAIQQAIRRKAITFVFEDTDLQLNP 131
L +AIRR+ T V D + +P
Sbjct 721 L---------------TEAIRRRPFTVVLFDEIEKAHP 743
> Hs21359886
Length=719
Score = 31.6 bits (70), Expect = 0.72, Method: Composition-based stats.
Identities = 17/64 (26%), Positives = 27/64 (42%), Gaps = 7/64 (10%)
Query 81 ASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFVFEDTDLQLNPTCAINITMN 140
A G CC DEF+++ +AI + + ++ I+ LN C+I N
Sbjct 437 ADQGVCCIDEFDKM-------AEADRTAIHEVMEQQTISIAKAGILTTLNARCSILAAAN 489
Query 141 PGYA 144
P Y
Sbjct 490 PAYG 493
> Hs7427519
Length=821
Score = 31.6 bits (70), Expect = 0.73, Method: Composition-based stats.
Identities = 30/104 (28%), Positives = 44/104 (42%), Gaps = 11/104 (10%)
Query 46 TTKDLAKAAGEPCLVFNCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQ 105
T+ + AAG V E + A L + A G CC DEF++ +DV V
Sbjct 419 TSGKASSAAGLTAAVVRDEESHEFVIEAGAL--MLADNGVCCIDEFDK--MDVRDQV--- 471
Query 106 ISAIQQAIRRKAITFVFEDTDLQLNPTCAINITMNP--GYAGRS 147
AI +A+ ++ I+ LN +I NP G+ RS
Sbjct 472 --AIHEAMEQQTISITKAGVKATLNARTSILAAANPISGHYDRS 513
> CE06149
Length=850
Score = 31.6 bits (70), Expect = 0.78, Method: Composition-based stats.
Identities = 27/92 (29%), Positives = 42/92 (45%), Gaps = 10/92 (10%)
Query 37 GPAGTGKTETTKDLAKAAGEPCLVFNCSEGLDAAAMAALLKG-------LAASGGWCC-- 87
GPAG GK+ + +A+ AG + N S+ A + +L+G L A C
Sbjct 337 GPAGLGKSTLARIVARQAGYSTIDVNASDARTVADLNKVLEGAVKTSRTLDADQRPACLI 396
Query 88 FDEFNRLQLDVLSIVALQISAI-QQAIRRKAI 118
DE + +D + + I A ++AIRR I
Sbjct 397 LDEIDGTPIDTIRHLVRCIQATGKKAIRRPII 428
> SPAC3F10.01
Length=649
Score = 30.8 bits (68), Expect = 1.2, Method: Composition-based stats.
Identities = 30/119 (25%), Positives = 50/119 (42%), Gaps = 23/119 (19%)
Query 37 GPAGTGKTETTKDLAKAAGEPCLVFNCSEGLDAAAMAA-----------LLKGLA---AS 82
G GT K++ K + + A P V+ +G AA + A L+G A A
Sbjct 301 GDPGTAKSQFLKFVERLA--PIAVYTSGKGSSAAGLTASIQRDSVTREFYLEGGAMVLAD 358
Query 83 GGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFVFEDTDLQLNPTCAINITMNP 141
GG C DEF++++ + AI +A+ ++ I+ LN ++ NP
Sbjct 359 GGIVCIDEFDKMRDE-------DRVAIHEAMEQQTISIAKAGITTILNSRTSVLAAANP 410
> ECU07g0490
Length=563
Score = 30.8 bits (68), Expect = 1.3, Method: Composition-based stats.
Identities = 23/114 (20%), Positives = 46/114 (40%), Gaps = 17/114 (14%)
Query 37 GPAGTGKTETTKDLAKAAGEPCLVFNCSEGLDAAAMAALLKG----------LAASGGWC 86
G +GTGK+ K ++ L A+ +G + A G C
Sbjct 269 GDSGTGKSHLLKTCSRLLSPAVLTNGVGTTQAGLTTCAVRQGREWVLEAGALVLADTGLC 328
Query 87 CFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFVFEDTDLQLNPTCAINITMN 140
C DEFN+L+++ + + + +A+ ++ ++ LN C++ +N
Sbjct 329 CIDEFNKLKVN-------EKNGLLEAMEQQTLSIAKAGIVSSLNTRCSVIAAIN 375
> HsM14149623
Length=543
Score = 30.8 bits (68), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 17/63 (26%), Positives = 27/63 (42%), Gaps = 7/63 (11%)
Query 81 ASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFVFEDTDLQLNPTCAINITMN 140
A G CC DEF+++ +AI + + ++ I+ LN C+I N
Sbjct 261 ADQGVCCIDEFDKM-------AEADRTAIHEVMEQQTISIAKAGILTTLNARCSILAAAN 313
Query 141 PGY 143
P Y
Sbjct 314 PAY 316
> SPBC25D12.03c
Length=760
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 32/122 (26%), Positives = 49/122 (40%), Gaps = 23/122 (18%)
Query 37 GPAGTGKTETTKDLAKAAGEPCLVFNCSEG-----LDAAAM------AALLKGLA---AS 82
G G K++ K ++K A P V+ G L AA M +L+G A A
Sbjct 403 GDPGVAKSQLLKYISKVA--PRGVYTTGRGSSGVGLTAAVMRDPVTDEMVLEGGALVLAD 460
Query 83 GGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFVFEDTDLQLNPTCAINITMNPG 142
G CC DEF+++ +AI + + ++ I+ LN +I NP
Sbjct 461 NGICCIDEFDKMDES-------DRTAIHEVMEQQTISISKAGITTTLNARTSILAAANPL 513
Query 143 YA 144
Y
Sbjct 514 YG 515
> At5g46280
Length=776
Score = 30.4 bits (67), Expect = 1.6, Method: Composition-based stats.
Identities = 27/100 (27%), Positives = 43/100 (43%), Gaps = 22/100 (22%)
Query 67 LDAAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFVFEDTD 126
L+A AM KG+ C DEF+++ D + AI + + ++ +T
Sbjct 383 LEAGAMVLADKGIV------CIDEFDKMN-DQDRV------AIHEVMEQQTVTIAKAGIH 429
Query 127 LQLNPTCAINITMNPGYA--GRSI-------LPDTLKALF 157
LN C++ NP Y RS+ LPD+L + F
Sbjct 430 ASLNARCSVVAAANPIYGTYDRSLTPTKNIGLPDSLLSRF 469
> At2g07690
Length=727
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 30/119 (25%), Positives = 49/119 (41%), Gaps = 23/119 (19%)
Query 37 GPAGTGKTETTKDLAKAAGEPCLVFNCSEGLDAAAMAA-----------LLKGLA---AS 82
G T K++ K + K A P V+ +G AA + A L+G A A
Sbjct 375 GDPSTAKSQFLKFVEKTA--PIAVYTSGKGSSAAGLTASVIRDSSTREFYLEGGAMVLAD 432
Query 83 GGWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFVFEDTDLQLNPTCAINITMNP 141
GG C DEF++++ + AI +A+ ++ I+ LN ++ NP
Sbjct 433 GGVVCIDEFDKMRPE-------DRVAIHEAMEQQTISIAKAGITTVLNSRTSVLAAANP 484
> ECU05g0780
Length=726
Score = 30.0 bits (66), Expect = 2.1, Method: Composition-based stats.
Identities = 30/119 (25%), Positives = 48/119 (40%), Gaps = 23/119 (19%)
Query 37 GPAGTGKTETTKDLAKAAGEPCLVFNCSEGLDAAAMAALLKGLAASG------------- 83
G GT K++ K +A P V+ + AA + A + +G
Sbjct 363 GDPGTAKSQFLKQ--ASAFLPRSVYTSGKSSSAAGLTASVVKDGETGEFTIEAGALMLSD 420
Query 84 -GWCCFDEFNRLQLDVLSIVALQISAIQQAIRRKAITFVFEDTDLQLNPTCAINITMNP 141
G CC DEF+++ V Q+S I +A+ ++ IT + LN +I NP
Sbjct 421 TGVCCIDEFDKMN------VKDQVS-IHEAMEQQTITISKAGINATLNARSSILAAANP 472
> YLR106c
Length=4910
Score = 30.0 bits (66), Expect = 2.4, Method: Composition-based stats.
Identities = 15/50 (30%), Positives = 28/50 (56%), Gaps = 0/50 (0%)
Query 20 YRTLMTALHFQYGGAPEGPAGTGKTETTKDLAKAAGEPCLVFNCSEGLDA 69
Y +++ A++ + GP+ +GKTET + LA G VF+ + +D+
Sbjct 2037 YESVLKAINNNWPLVLVGPSNSGKTETIRFLASILGPRVDVFSMNSDIDS 2086
Score = 29.6 bits (65), Expect = 2.4, Method: Composition-based stats.
Identities = 39/167 (23%), Positives = 61/167 (36%), Gaps = 28/167 (16%)
Query 11 VVTPLTDRCYRTLMTALHFQ-YGGAPEGPAGTGKTETTKDLAKAAGEPCLVFNCSEGLD- 68
++TP ++ L+ A + + +GP +GKT K LA G + N E D
Sbjct 1056 IITPFVEKNMMNLVRATSGKRFPVLIQGPTSSGKTSMIKYLADITGHKFVRINNHEHTDL 1115
Query 69 ---------------AAAMAALLKGLAASGGWCCFDEFNRLQLDVLSIVALQISAIQQAI 113
+ L++ L G W DE N DVL A+ + +
Sbjct 1116 QEYLGTYVTDDTGKLSFKEGVLVEAL-RKGYWIVLDELNLAPTDVL-------EALNRLL 1167
Query 114 RRKAITFVFEDTD-LQLNPTCAINITMNPG--YAGRSILPDTLKALF 157
F+ E + + +P + T NP Y GR IL + F
Sbjct 1168 DDNRELFIPETQEVVHPHPDFLLFATQNPPGIYGGRKILSRAFRNRF 1214
> At4g04180
Length=536
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 13/24 (54%), Positives = 15/24 (62%), Gaps = 0/24 (0%)
Query 36 EGPAGTGKTETTKDLAKAAGEPCL 59
EGP GTGKT + +A AG P L
Sbjct 294 EGPPGTGKTSCARVIANQAGIPLL 317
> Hs7706294
Length=438
Score = 29.6 bits (65), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 20/56 (35%), Positives = 28/56 (50%), Gaps = 2/56 (3%)
Query 42 GKTETTKDLAKAAGEPCLVFNCSEGLDAAAMAALLKGLAASGGWCCFDEFNRLQLD 97
G +TT ++ +A C + + G AAA A L A GG C DE + L+LD
Sbjct 372 GSLKTTNEILDSASHDCPLVTQTYG--AAAGKAYTDDLGAVGGACLEDEASALRLD 425
> YDR258c
Length=811
Score = 29.6 bits (65), Expect = 2.7, Method: Composition-based stats.
Identities = 29/90 (32%), Positives = 41/90 (45%), Gaps = 21/90 (23%)
Query 37 GPAGTGKTETTKDLAKAAGEP---CLVFNCSEGLDAAAMAALLKG-----LAASGGW--- 85
GP GTGKTE TK LA+ + + F+ SE + ++ L+ L+ SGG
Sbjct 541 GPTGTGKTELTKALAEFLFDDESNVIRFDMSEFQEKHTVSRLIGAPPGYVLSESGGQLTE 600
Query 86 ---------CCFDEFNRLQLDVLSIVALQI 106
FDEF + DV S + LQ+
Sbjct 601 AVRRKPYAVVLFDEFEKAHPDV-SKLLLQV 629
> At1g31260
Length=355
Score = 29.6 bits (65), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 20/84 (23%), Positives = 38/84 (45%), Gaps = 6/84 (7%)
Query 81 ASGGWCCFDEFNRLQLDVLSIVALQISAIQQA----IRRKAITFVFEDTDLQLNPTCAIN 136
+ + C D+ L L +LSI ++ I+++ R F E + + + A
Sbjct 36 SKSNYSCIDKNKALDLKLLSIFSILITSLIGVCLPFFARSIPAFQPEKSHFLIVKSFASG 95
Query 137 ITMNPGYAGRSILPDTLKALFRPC 160
I ++ G+ +LPD+ + L PC
Sbjct 96 IILSTGF--MHVLPDSFEMLSSPC 117
Lambda K H
0.324 0.137 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2281134618
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40