bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2607_orf1
Length=134
Score E
Sequences producing significant alignments: (Bits) Value
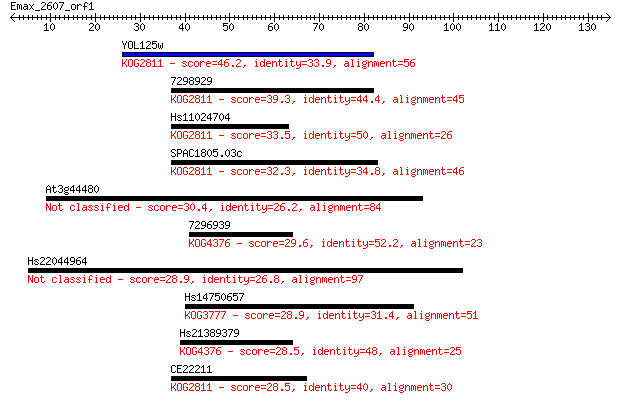
YOL125w 46.2 2e-05
7298929 39.3 0.002
Hs11024704 33.5 0.11
SPAC1805.03c 32.3 0.23
At3g44480 30.4 1.1
7296939 29.6 1.6
Hs22044964 28.9 2.7
Hs14750657 28.9 2.7
Hs21389379 28.5 3.4
CE22211 28.5 4.0
> YOL125w
Length=476
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 19/56 (33%), Positives = 34/56 (60%), Gaps = 0/56 (0%)
Query 26 SRNRISSGRHGERIDCPMDNSHSIYALRLQAHLKKCTKARDIAFSHCLPFMRPGVN 81
++N + + ER+ CP+D +H+++A +L+ HLKKC K + + P+ PG N
Sbjct 60 NKNGSEAEKERERVPCPLDPNHTVWADQLKKHLKKCNKTKLSHLNDDKPYYEPGYN 115
> 7298929
Length=434
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/45 (44%), Positives = 28/45 (62%), Gaps = 3/45 (6%)
Query 37 ERIDCPMDNSHSIYALRLQAHLKKCTKARDIAFSHCLPFMRPGVN 81
ERI CP+D+ H+++ +L HL C ARD S LP++ GVN
Sbjct 60 ERIPCPLDHKHTVFKRKLAKHLTICN-ARDQESS--LPYIVKGVN 101
> Hs11024704
Length=481
Score = 33.5 bits (75), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 13/26 (50%), Positives = 18/26 (69%), Gaps = 0/26 (0%)
Query 37 ERIDCPMDNSHSIYALRLQAHLKKCT 62
+RI CP+D H++Y +L HLKKC
Sbjct 55 KRILCPLDPKHTVYEDQLAKHLKKCN 80
> SPAC1805.03c
Length=407
Score = 32.3 bits (72), Expect = 0.23, Method: Composition-based stats.
Identities = 16/46 (34%), Positives = 28/46 (60%), Gaps = 3/46 (6%)
Query 37 ERIDCPMDNSHSIYALRLQAHLKKCTKARDIAFSHCLPFMRPGVNL 82
++I CP D+ H+I RL+ HLK+C AR + + P+ + +N+
Sbjct 53 KQIPCPYDHKHTIVRHRLEYHLKRCN-ARPVERTD--PYYKKDINI 95
> At3g44480
Length=1220
Score = 30.4 bits (67), Expect = 1.1, Method: Composition-based stats.
Identities = 22/89 (24%), Positives = 39/89 (43%), Gaps = 14/89 (15%)
Query 9 SATXCGAEAAVPQ-DEALSRNRISSGRHGERIDCPMDNSHSIYALRLQAHLKKCTK---- 63
S C ++PQ ++L + + ER+DC +N ++ + KC K
Sbjct 972 SLNNCNNLVSLPQLSDSLDYIYADNCKSLERLDCCFNNPE------IRLYFPKCFKLNQE 1025
Query 64 ARDIAFSHCLPFMRPGVNLPPQQHCYLEQ 92
ARD+ C+ M PG +P C++ +
Sbjct 1026 ARDLIMHTCIDAMFPGTQVPA---CFIHR 1051
> 7296939
Length=93
Score = 29.6 bits (65), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 12/23 (52%), Positives = 13/23 (56%), Gaps = 0/23 (0%)
Query 41 CPMDNSHSIYALRLQAHLKKCTK 63
CP D SH I R+ HL KC K
Sbjct 9 CPYDKSHRILLFRMPKHLIKCEK 31
> Hs22044964
Length=2192
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 26/107 (24%), Positives = 44/107 (41%), Gaps = 13/107 (12%)
Query 5 PDATSATXCGAEAAVPQDEALSRNRISSGRHGERI----------DCPMDNSHSIYALRL 54
P+ + AEA + + + S R+ G R+ D P + S +I+ +
Sbjct 1688 PERNRVSSYSAEALIGKTSSNSEQRMGISIQGSRVSDQLEMRSYLDVPRNKSLAIH--NM 1745
Query 55 QAHLKKCTKARDIAFSHCLPFMRPGVNLPPQQHCYLEQKQQQEHGHP 101
Q + T A DI S C F G + PQ + ++ + E G+P
Sbjct 1746 QGRVDH-TVASDIRLSDCQTFKPSGASQQPQSNFEVQSSRNNEIGNP 1791
> Hs14750657
Length=719
Score = 28.9 bits (63), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 16/51 (31%), Positives = 29/51 (56%), Gaps = 3/51 (5%)
Query 40 DCPMDNSHSIYALRLQAHLKKCTKARDIAFSHCLPFMRPGVNLPPQQHCYL 90
DCP DN H I+ L+L+ + ++A++ C P ++ ++ PP+ HC
Sbjct 86 DCP-DNPH-IW-LQLEGPKENASRAKEYLKGLCSPELQDEIHYPPKLHCIF 133
> Hs21389379
Length=167
Score = 28.5 bits (62), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 12/25 (48%), Positives = 14/25 (56%), Gaps = 0/25 (0%)
Query 39 IDCPMDNSHSIYALRLQAHLKKCTK 63
+ CP D +H I A R HL KC K
Sbjct 15 LQCPYDKNHQIRACRFPYHLIKCRK 39
> CE22211
Length=390
Score = 28.5 bits (62), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 12/31 (38%), Positives = 19/31 (61%), Gaps = 1/31 (3%)
Query 37 ERIDCPMDNSHSIYALRLQAHLKKC-TKARD 66
+R+ CP D H+I L+ HL +C ++ RD
Sbjct 50 DRMVCPNDGKHTILKADLEIHLTRCNSRIRD 80
Lambda K H
0.315 0.129 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1393086308
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40