bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2643_orf1
Length=121
Score E
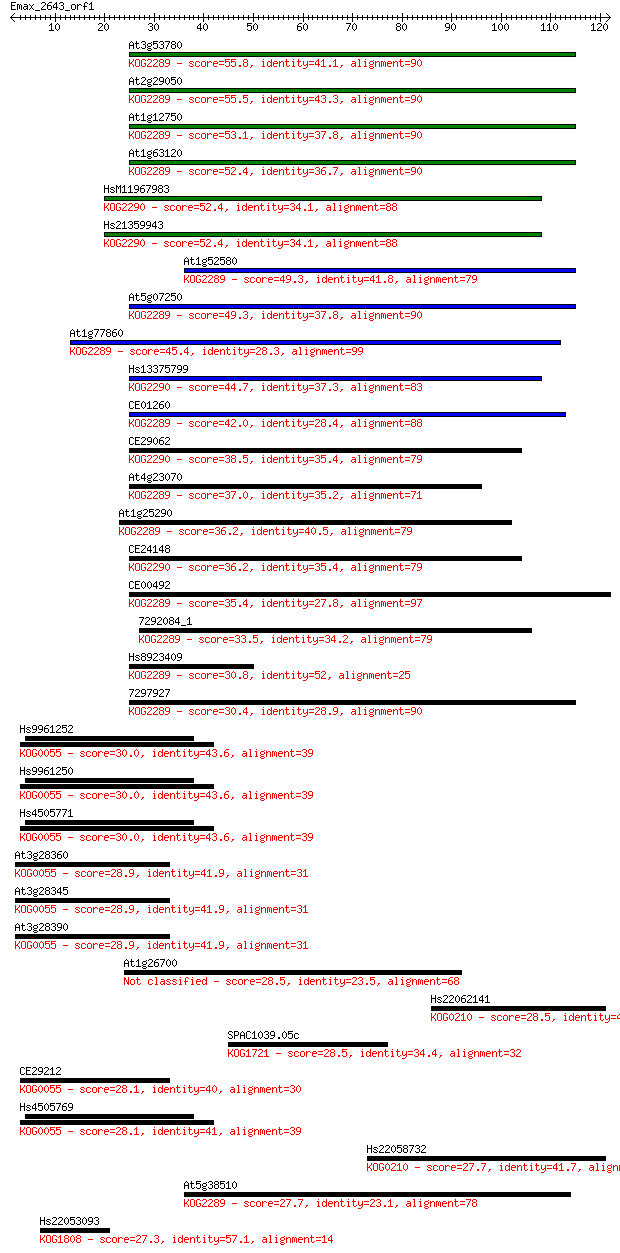
Sequences producing significant alignments: (Bits) Value
At3g53780 55.8 2e-08
At2g29050 55.5 3e-08
At1g12750 53.1 1e-07
At1g63120 52.4 2e-07
HsM11967983 52.4 2e-07
Hs21359943 52.4 2e-07
At1g52580 49.3 2e-06
At5g07250 49.3 2e-06
At1g77860 45.4 2e-05
Hs13375799 44.7 5e-05
CE01260 42.0 2e-04
CE29062 38.5 0.003
At4g23070 37.0 0.010
At1g25290 36.2 0.015
CE24148 36.2 0.017
CE00492 35.4 0.025
7292084_1 33.5 0.11
Hs8923409 30.8 0.66
7297927 30.4 0.84
Hs9961252 30.0 1.0
Hs9961250 30.0 1.0
Hs4505771 30.0 1.0
At3g28360 28.9 2.2
At3g28345 28.9 2.5
At3g28390 28.9 2.8
At1g26700 28.5 3.0
Hs22062141 28.5 3.3
SPAC1039.05c 28.5 3.6
CE29212 28.1 4.1
Hs4505769 28.1 4.6
Hs22058732 27.7 5.3
At5g38510 27.7 5.8
Hs22053093 27.3 6.3
> At3g53780
Length=361
Score = 55.8 bits (133), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 37/90 (41%), Positives = 52/90 (57%), Gaps = 4/90 (4%)
Query 25 LLSAVFDPCTTTVGSSGALYGLLGGLVPFAVENWDFLHYPWCVLAIVIVVIATAQLNSMG 84
+LSA+F +VG+SGA++GLLGG++ NW Y V+ IV +V+ A +G
Sbjct 158 ILSALFLRSNISVGASGAVFGLLGGMLSEIFINWTI--YSNKVVTIVTLVLIVAVNLGLG 215
Query 85 GFSAVDNFAHLGGCLGGLLFGFATIYTLRP 114
VDNFAH+GG G L GF + +RP
Sbjct 216 VLPGVDNFAHIGGFATGFLLGF--VLLIRP 243
> At2g29050
Length=372
Score = 55.5 bits (132), Expect = 3e-08, Method: Composition-based stats.
Identities = 39/118 (33%), Positives = 56/118 (47%), Gaps = 32/118 (27%)
Query 25 LLSAVFDPCTTTVGSSGALYGLLGGLVPFAVENWD------------------------- 59
LLS++F+ +VG+SGAL+GLLG ++ + NW
Sbjct 207 LLSSLFNRAGISVGASGALFGLLGAMLSELLTNWTIYANKVAKSSLVKQAALSMNDVSIM 266
Query 60 ---FLHYPWCVLAIVIVVIATAQLNSMGGFSAVDNFAHLGGCLGGLLFGFATIYTLRP 114
FLH + L +I +IA ++G VDNFAHLGG G L GF ++ +RP
Sbjct 267 SLVFLHLQFAALLTLIFIIAINL--AVGILPHVDNFAHLGGFTSGFLLGF--VFLIRP 320
> At1g12750
Length=307
Score = 53.1 bits (126), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 34/90 (37%), Positives = 50/90 (55%), Gaps = 4/90 (4%)
Query 25 LLSAVFDPCTTTVGSSGALYGLLGGLVPFAVENWDFLHYPWCVLAIVIVVIATAQLNSMG 84
+LSA+F + +VG+SGAL GL+G ++ + NW C L + +IA ++G
Sbjct 149 ILSALFLQKSISVGASGALLGLMGAMLSELLTNWTIYKSKLCALLSFLFIIAINL--AIG 206
Query 85 GFSAVDNFAHLGGCLGGLLFGFATIYTLRP 114
VDNFAH+GG L G GF I ++P
Sbjct 207 LLPWVDNFAHIGGLLTGFCLGF--ILLMQP 234
> At1g63120
Length=317
Score = 52.4 bits (124), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 33/90 (36%), Positives = 51/90 (56%), Gaps = 4/90 (4%)
Query 25 LLSAVFDPCTTTVGSSGALYGLLGGLVPFAVENWDFLHYPWCVLAIVIVVIATAQLNSMG 84
+LS++F + +VG+SGAL+GLLG ++ + NW L ++ +IA ++G
Sbjct 162 ILSSLFLQESISVGASGALFGLLGAMLSELLTNWTIYANKAAALITLLFIIAINL--ALG 219
Query 85 GFSAVDNFAHLGGCLGGLLFGFATIYTLRP 114
VDNFAH+GG L G GF + +RP
Sbjct 220 MLPRVDNFAHIGGFLTGFCLGFVLL--VRP 247
> HsM11967983
Length=855
Score = 52.4 bits (124), Expect = 2e-07, Method: Composition-based stats.
Identities = 30/96 (31%), Positives = 47/96 (48%), Gaps = 10/96 (10%)
Query 20 VDVVYLLS--------AVFDPCTTTVGSSGALYGLLGGLVPFAVENWDFLHYPWCVLAIV 71
+ ++YLLS A+F P VG +G+ +G+L L ++W L PW +
Sbjct 692 IAIIYLLSGVTGNLASAIFLPYRAEVGPAGSQFGILACLFVELFQSWQILARPWRAFFKL 751
Query 72 IVVIATAQLNSMGGFSAVDNFAHLGGCLGGLLFGFA 107
+ V+ L + G +DNFAH+ G + GL FA
Sbjct 752 LAVVLF--LFTFGLLPWIDNFAHISGFISGLFLSFA 785
> Hs21359943
Length=855
Score = 52.4 bits (124), Expect = 2e-07, Method: Composition-based stats.
Identities = 30/96 (31%), Positives = 47/96 (48%), Gaps = 10/96 (10%)
Query 20 VDVVYLLS--------AVFDPCTTTVGSSGALYGLLGGLVPFAVENWDFLHYPWCVLAIV 71
+ ++YLLS A+F P VG +G+ +G+L L ++W L PW +
Sbjct 692 IAIIYLLSGVTGNLASAIFLPYRAEVGPAGSQFGILACLFVELFQSWQILARPWRAFFKL 751
Query 72 IVVIATAQLNSMGGFSAVDNFAHLGGCLGGLLFGFA 107
+ V+ L + G +DNFAH+ G + GL FA
Sbjct 752 LAVVLF--LFTFGLLPWIDNFAHISGFISGLFLSFA 785
> At1g52580
Length=309
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/80 (41%), Positives = 44/80 (55%), Gaps = 6/80 (7%)
Query 36 TVGSSGALYGLLGGLVPFAVENWDFLHYPWCVLAIVIVVIATAQLN-SMGGFSAVDNFAH 94
+VG+SGAL+GLLG ++ + NW L +I++I LN S+G VDN AH
Sbjct 171 SVGASGALFGLLGAMLSELITNWTIYENKCTALMTLILIIV---LNLSVGFLPRVDNSAH 227
Query 95 LGGCLGGLLFGFATIYTLRP 114
GG L G GF + LRP
Sbjct 228 FGGFLAGFFLGFVLL--LRP 245
> At5g07250
Length=346
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 34/90 (37%), Positives = 49/90 (54%), Gaps = 4/90 (4%)
Query 25 LLSAVFDPCTTTVGSSGALYGLLGGLVPFAVENWDFLHYPWCVLAIVIVVIATAQLNSMG 84
+LS++F + +VG+SGAL+GLLG ++ NW L ++ VI ++G
Sbjct 186 VLSSLFIRNSISVGASGALFGLLGSMLSELFTNWTIYSNKIAALLTLLFVILINL--AIG 243
Query 85 GFSAVDNFAHLGGCLGGLLFGFATIYTLRP 114
VDNFAH+GG + G L GF I RP
Sbjct 244 ILPHVDNFAHVGGFVTGFLLGF--ILLARP 271
> At1g77860
Length=319
Score = 45.4 bits (106), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 28/107 (26%), Positives = 50/107 (46%), Gaps = 10/107 (9%)
Query 13 RGCGSAVVDVVYLLSAV--------FDPCTTTVGSSGALYGLLGGLVPFAVENWDFLHYP 64
+ G + V+Y LS + F ++ S A +GL+G ++ +NW+ +
Sbjct 154 QQFGPLRIAVIYFLSGIMGSLFAVLFVRNIPSISSGAAFFGLIGAMLSALAKNWNLYNSK 213
Query 65 WCVLAIVIVVIATAQLNSMGGFSAVDNFAHLGGCLGGLLFGFATIYT 111
LAI+ + L +G +DNFA++GG + G L GF ++
Sbjct 214 ISALAIIFTIFTVNFL--IGFLPFIDNFANIGGFISGFLLGFVLLFK 258
> Hs13375799
Length=619
Score = 44.7 bits (104), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 31/87 (35%), Positives = 41/87 (47%), Gaps = 10/87 (11%)
Query 25 LLSAVFDPCTTTVGSSGALYGLLGGLVPFAVENWDFLHYPWCVL----AIVIVVIATAQL 80
L SA+F P VG +G+ +GLL L ++W L PW AIV+ +
Sbjct 469 LASAIFLPYRAEVGPAGSQFGLLACLFVELFQSWPLLERPWKAFLNLSAIVLFLFIC--- 525
Query 81 NSMGGFSAVDNFAHLGGCLGGLLFGFA 107
G +DN AH+ G L GLL FA
Sbjct 526 ---GLLPWIDNIAHIFGFLSGLLLAFA 549
> CE01260
Length=419
Score = 42.0 bits (97), Expect = 2e-04, Method: Composition-based stats.
Identities = 25/94 (26%), Positives = 42/94 (44%), Gaps = 6/94 (6%)
Query 25 LLSAVFDPCTTTVGSSGALYGLLGGLVPFAVENWDFLHYPWCVLAIVIVVI------ATA 78
LL DP + VG+S +Y L+ V + NW + W + ++ V I A
Sbjct 281 LLQYAIDPNSLLVGASAGVYALIFAHVANVILNWHEMPLRWIRVLVLFVFIFLDFGGAIH 340
Query 79 QLNSMGGFSAVDNFAHLGGCLGGLLFGFATIYTL 112
+ +V + AH+ G + GL FG+ +Y +
Sbjct 341 RRFYTNDCDSVSHLAHIAGAVTGLFFGYVVLYNV 374
> CE29062
Length=727
Score = 38.5 bits (88), Expect = 0.003, Method: Composition-based stats.
Identities = 28/80 (35%), Positives = 41/80 (51%), Gaps = 4/80 (5%)
Query 25 LLSAVFDPCTTTVGSSGALYGLLGGLVPFAVENWDFL-HYPWCVLAIVIVVIATAQLNSM 83
L SA+F P VG S A G+L ++ +N + + W A+V +I T + +
Sbjct 504 LASAIFVPYNPAVGPSSAQCGILAAVIVECCDNRRIIKEFKW---ALVQHLIVTLLVLCI 560
Query 84 GGFSAVDNFAHLGGCLGGLL 103
G VDN+AHL G + GLL
Sbjct 561 GFIPWVDNWAHLFGTIFGLL 580
> At4g23070
Length=313
Score = 37.0 bits (84), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 25/71 (35%), Positives = 40/71 (56%), Gaps = 2/71 (2%)
Query 25 LLSAVFDPCTTTVGSSGALYGLLGGLVPFAVENWDFLHYPWCVLAIVIVVIATAQLNSMG 84
+LS +F +VG+S AL+GLLG ++ + NW Y +AIV++++ +G
Sbjct 156 ILSCLFLEDAISVGASSALFGLLGAMLSELLINWT--TYDNKGVAIVMLLVIVGVNLGLG 213
Query 85 GFSAVDNFAHL 95
VDNFAH+
Sbjct 214 TLPPVDNFAHI 224
> At1g25290
Length=369
Score = 36.2 bits (82), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 32/95 (33%), Positives = 47/95 (49%), Gaps = 19/95 (20%)
Query 23 VYLLSAVFDPC--------------TTTVGSSGALYGLLGGLVPFAVENWDFLHYPWCVL 68
VYL SAV P +VG+SGA++GL+G + F + + + L
Sbjct 235 VYLTSAVAKPILRVLGSAMSYWFNKAPSVGASGAIFGLVGSVAVFVIRHKQMVRGGNEDL 294
Query 69 AIVIVVIATAQLN-SMGGFS-AVDNFAHLGGCLGG 101
+ +IA LN +MG S +DN+ H+GG LGG
Sbjct 295 MQIAQIIA---LNMAMGLMSRRIDNWGHIGGLLGG 326
> CE24148
Length=851
Score = 36.2 bits (82), Expect = 0.017, Method: Composition-based stats.
Identities = 28/82 (34%), Positives = 42/82 (51%), Gaps = 6/82 (7%)
Query 25 LLSAVFDPCTTTVGSSGALYGLLGGLVPFAVENWDFLHY--PWCV-LAIVIVVIATAQLN 81
L SA+F P TVG S A G+ +V VE W F H P+ + + +I T +
Sbjct 627 LASAIFVPYYPTVGPSSAQCGVFSSVV---VELWHFRHLLDPFELKFQSIAHLIVTLLVL 683
Query 82 SMGGFSAVDNFAHLGGCLGGLL 103
+G +DN++HL G + GL+
Sbjct 684 CIGLIPWIDNWSHLFGTIFGLI 705
> CE00492
Length=397
Score = 35.4 bits (80), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 27/103 (26%), Positives = 43/103 (41%), Gaps = 6/103 (5%)
Query 25 LLSAVFDPCTTTVGSSGALYGLLGGLVPFAVENWDFLHYPWCVLAIVIV------VIATA 78
+LS DP G + + L+ + N+ + C L I+IV V+A
Sbjct 182 ILSLALDPTVFLCGGAAGSFSLIASHITTIATNFKEMENATCRLPILIVFAALDYVLAVY 241
Query 79 QLNSMGGFSAVDNFAHLGGCLGGLLFGFATIYTLRPIRRFSLS 121
Q V + HLGG + G+LF F +P R +++S
Sbjct 242 QRFFAPRIDKVSMYGHLGGLVAGILFTFILFRGSKPSRFYTVS 284
> 7292084_1
Length=356
Score = 33.5 bits (75), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 42/102 (41%), Gaps = 23/102 (22%)
Query 27 SAVFDPCTTTVGSSGALYGLLGGLVPFAVENWDFLHYPWCVLAIVIVVIA---------- 76
+++FDP VG+SG +Y LL + + N+ + Y L ++V ++
Sbjct 167 TSIFDPDVFLVGASGGVYALLAAHLANVLLNYHQMRYGVIKLLHILVFVSFDFGFAIYAR 226
Query 77 ----TAQLNSMGGFSAVDN---------FAHLGGCLGGLLFG 105
QL S F A+D AHL G + GL G
Sbjct 227 YAGDELQLGSSSEFLAIDQAETAGAVSYVAHLAGAIAGLTIG 268
> Hs8923409
Length=292
Score = 30.8 bits (68), Expect = 0.66, Method: Compositional matrix adjust.
Identities = 13/25 (52%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 25 LLSAVFDPCTTTVGSSGALYGLLGG 49
L S++FDP VG+SG +Y L+GG
Sbjct 161 LASSIFDPLRYLVGASGGVYALMGG 185
> 7297927
Length=263
Score = 30.4 bits (67), Expect = 0.84, Method: Compositional matrix adjust.
Identities = 26/97 (26%), Positives = 46/97 (47%), Gaps = 7/97 (7%)
Query 25 LLSAVFDPCTTTVGSSGALYGLLGGLVPFAVENWDFLHYPWCVLAIVIVVIATAQLNSMG 84
L +A P +G+S +Y +LG VP V N+ L + + +A +++++ + +
Sbjct 159 LANAWLQPHLHLMGASAGVYAMLGSHVPHLVLNFSQLSHRFARIASLLILLLSDVGFTTY 218
Query 85 GFSAVDNF-------AHLGGCLGGLLFGFATIYTLRP 114
F N AH+GG + G+L GF L+P
Sbjct 219 HFCHNHNRNPRTSLEAHIGGGVAGILCGFIVYRRLQP 255
> Hs9961252
Length=1232
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 2/34 (5%)
Query 4 GDSLCLLGGRGCGSAVVDVVYLLSAVFDPCTTTV 37
G +L L+G GCG + VV LL +DP TV
Sbjct 1015 GQTLALVGSSGCGKST--VVQLLERFYDPLAGTV 1046
Score = 28.1 bits (61), Expect = 4.7, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 21/39 (53%), Gaps = 2/39 (5%)
Query 3 SGDSLCLLGGRGCGSAVVDVVYLLSAVFDPCTTTVGSSG 41
SG ++ L+G GCG + V L+ ++DP T+ G
Sbjct 421 SGQTVALVGSSGCGKST--TVQLIQRLYDPDEGTINIDG 457
> Hs9961250
Length=1286
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 2/34 (5%)
Query 4 GDSLCLLGGRGCGSAVVDVVYLLSAVFDPCTTTV 37
G +L L+G GCG + VV LL +DP TV
Sbjct 1062 GQTLALVGSSGCGKST--VVQLLERFYDPLAGTV 1093
Score = 28.1 bits (61), Expect = 4.7, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 21/39 (53%), Gaps = 2/39 (5%)
Query 3 SGDSLCLLGGRGCGSAVVDVVYLLSAVFDPCTTTVGSSG 41
SG ++ L+G GCG + V L+ ++DP T+ G
Sbjct 421 SGQTVALVGSSGCGKST--TVQLIQRLYDPDEGTINIDG 457
> Hs4505771
Length=1279
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 2/34 (5%)
Query 4 GDSLCLLGGRGCGSAVVDVVYLLSAVFDPCTTTV 37
G +L L+G GCG + VV LL +DP TV
Sbjct 1062 GQTLALVGSSGCGKST--VVQLLERFYDPLAGTV 1093
Score = 28.1 bits (61), Expect = 4.7, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 21/39 (53%), Gaps = 2/39 (5%)
Query 3 SGDSLCLLGGRGCGSAVVDVVYLLSAVFDPCTTTVGSSG 41
SG ++ L+G GCG + V L+ ++DP T+ G
Sbjct 421 SGQTVALVGSSGCGKST--TVQLIQRLYDPDEGTINIDG 457
> At3g28360
Length=1158
Score = 28.9 bits (63), Expect = 2.2, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 19/31 (61%), Gaps = 2/31 (6%)
Query 2 PSGDSLCLLGGRGCGSAVVDVVYLLSAVFDP 32
PSG ++ L+GG G G + V+ LL +DP
Sbjct 302 PSGKTVALVGGSGSGKST--VISLLQRFYDP 330
> At3g28345
Length=1240
Score = 28.9 bits (63), Expect = 2.5, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 19/31 (61%), Gaps = 2/31 (6%)
Query 2 PSGDSLCLLGGRGCGSAVVDVVYLLSAVFDP 32
PSG ++ L+GG G G + V+ LL +DP
Sbjct 385 PSGKTVALVGGSGSGKST--VISLLQRFYDP 413
> At3g28390
Length=1225
Score = 28.9 bits (63), Expect = 2.8, Method: Composition-based stats.
Identities = 13/31 (41%), Positives = 19/31 (61%), Gaps = 2/31 (6%)
Query 2 PSGDSLCLLGGRGCGSAVVDVVYLLSAVFDP 32
PSG ++ L+GG G G + V+ LL +DP
Sbjct 373 PSGKTVALVGGSGSGKST--VISLLQRFYDP 401
> At1g26700
Length=610
Score = 28.5 bits (62), Expect = 3.0, Method: Composition-based stats.
Identities = 16/68 (23%), Positives = 38/68 (55%), Gaps = 1/68 (1%)
Query 24 YLLSAVFDPCTTTVGSSGALYGLLGGLVPFAVENWDFLHYPWCVLAIVIVVIATAQLNSM 83
Y++ ++ + VG SG L+G + G + F ++ + L++ ++ I +V++ A+L +
Sbjct 327 YMIRSMEEEFQKIVGVSGPLWGFVVGFMLFNIKGSN-LYFWLAIIPITLVLLVGAKLQHV 385
Query 84 GGFSAVDN 91
A++N
Sbjct 386 IATLALEN 393
> Hs22062141
Length=875
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 16/35 (45%), Positives = 23/35 (65%), Gaps = 2/35 (5%)
Query 86 FSAVDNFAHLGGCLGGLLFGFATIYTLRPIRRFSL 120
FS+V FA + G L+ G++TIYT+ P+ FSL
Sbjct 688 FSSVFYFASVPLYQGFLIIGYSTIYTMFPV--FSL 720
> SPAC1039.05c
Length=781
Score = 28.5 bits (62), Expect = 3.6, Method: Composition-based stats.
Identities = 11/32 (34%), Positives = 18/32 (56%), Gaps = 0/32 (0%)
Query 45 GLLGGLVPFAVENWDFLHYPWCVLAIVIVVIA 76
LGG + + V+N LH PWC+ +++ A
Sbjct 670 AFLGGDMEYDVQNETGLHRPWCLYVSTLILWA 701
> CE29212
Length=1265
Score = 28.1 bits (61), Expect = 4.1, Method: Composition-based stats.
Identities = 12/30 (40%), Positives = 18/30 (60%), Gaps = 2/30 (6%)
Query 3 SGDSLCLLGGRGCGSAVVDVVYLLSAVFDP 32
+GD + L+G GCG + +V LL +DP
Sbjct 419 AGDKIALVGSSGCGKST--IVNLLQRFYDP 446
> Hs4505769
Length=1279
Score = 28.1 bits (61), Expect = 4.6, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 18/34 (52%), Gaps = 2/34 (5%)
Query 4 GDSLCLLGGRGCGSAVVDVVYLLSAVFDPCTTTV 37
G +L L+G GCG + VV LL +DP V
Sbjct 1062 GQTLALVGSSGCGKST--VVQLLERFYDPLAGKV 1093
Score = 27.7 bits (60), Expect = 5.0, Method: Composition-based stats.
Identities = 13/39 (33%), Positives = 20/39 (51%), Gaps = 2/39 (5%)
Query 3 SGDSLCLLGGRGCGSAVVDVVYLLSAVFDPCTTTVGSSG 41
SG ++ L+G GCG + V L+ ++DP V G
Sbjct 418 SGQTVALVGNSGCGKST--TVQLMQRLYDPTEGMVSVDG 454
> Hs22058732
Length=355
Score = 27.7 bits (60), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 29/48 (60%), Gaps = 6/48 (12%)
Query 73 VVIATAQLNSMGGFSAVDNFAHLGGCLGGLLFGFATIYTLRPIRRFSL 120
++I+T Q FS+V FA + G L+ G+ATIYT+ P+ FSL
Sbjct 148 LIISTMQ----AVFSSVFYFASVPLYQGFLMVGYATIYTMFPV--FSL 189
> At5g38510
Length=434
Score = 27.7 bits (60), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 18/78 (23%), Positives = 36/78 (46%), Gaps = 0/78 (0%)
Query 36 TVGSSGALYGLLGGLVPFAVENWDFLHYPWCVLAIVIVVIATAQLNSMGGFSAVDNFAHL 95
TVG +G + L+G + +N + + +I T + F +D++ +L
Sbjct 293 TVGGTGPAFALIGAWLVDQNQNKEMIKSNEYEDLFQKAIIMTGFGLILSHFGPIDDWTNL 352
Query 96 GGCLGGLLFGFATIYTLR 113
G + G+++GF T L+
Sbjct 353 GALIAGIVYGFFTCPVLQ 370
> Hs22053093
Length=1524
Score = 27.3 bits (59), Expect = 6.3, Method: Composition-based stats.
Identities = 8/14 (57%), Positives = 12/14 (85%), Gaps = 0/14 (0%)
Query 7 LCLLGGRGCGSAVV 20
+CL+GG+GCG V+
Sbjct 62 ICLIGGKGCGKTVI 75
Lambda K H
0.327 0.146 0.473
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1198419392
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40