bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
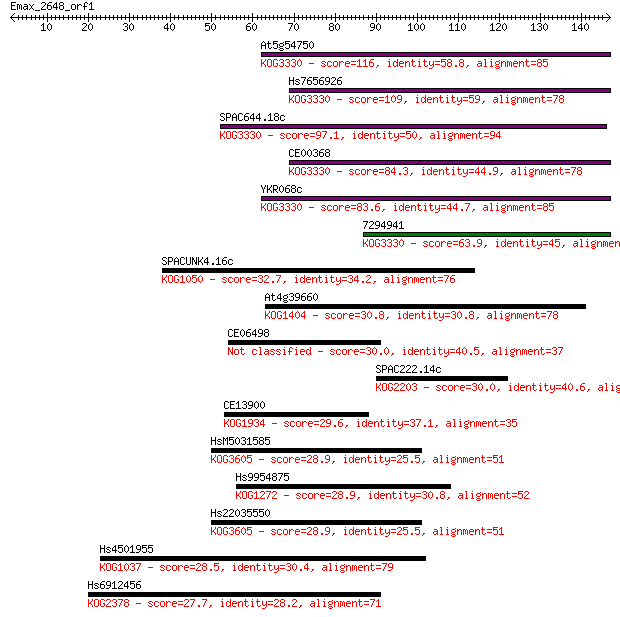
Query= Emax_2648_orf1
Length=146
Score E
Sequences producing significant alignments: (Bits) Value
At5g54750 116 2e-26
Hs7656926 109 2e-24
SPAC644.18c 97.1 1e-20
CE00368 84.3 8e-17
YKR068c 83.6 1e-16
7294941 63.9 1e-10
SPACUNK4.16c 32.7 0.23
At4g39660 30.8 1.1
CE06498 30.0 1.4
SPAC222.14c 30.0 1.7
CE13900 29.6 2.1
HsM5031585 28.9 3.3
Hs9954875 28.9 3.5
Hs22035550 28.9 3.7
Hs4501955 28.5 4.4
Hs6912456 27.7 7.8
> At5g54750
Length=186
Score = 116 bits (290), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 50/85 (58%), Positives = 68/85 (80%), Gaps = 0/85 (0%)
Query 62 EAISHKIEKANSELLCLSYGSLVSQLLKDFEQVEAVNAQLEKMGYNIGVRLVDEFLAKAN 121
+AI I++ N+EL L+YG++V QLL D E+VE VN QL++MGYNIG+RL+DEFLAK+
Sbjct 10 DAIFSSIDRVNAELFTLTYGAIVRQLLTDLEEVEEVNKQLDQMGYNIGIRLIDEFLAKSG 69
Query 122 IGACESFRETAEVIAKLGLRMFLGV 146
+ C F+ETAE+IAK+G +MFLGV
Sbjct 70 VSRCVDFKETAEMIAKVGFKMFLGV 94
> Hs7656926
Length=180
Score = 109 bits (273), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 46/78 (58%), Positives = 65/78 (83%), Gaps = 0/78 (0%)
Query 69 EKANSELLCLSYGSLVSQLLKDFEQVEAVNAQLEKMGYNIGVRLVDEFLAKANIGACESF 128
+K +SEL L+YG+LV+QL KD+E E VN QL+KMG+NIGVRL+++FLA++N+G C F
Sbjct 12 KKMSSELFTLTYGALVTQLCKDYENDEDVNKQLDKMGFNIGVRLIEDFLARSNVGRCHDF 71
Query 129 RETAEVIAKLGLRMFLGV 146
RETA+VIAK+ +M+LG+
Sbjct 72 RETADVIAKVAFKMYLGI 89
> SPAC644.18c
Length=183
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 47/94 (50%), Positives = 64/94 (68%), Gaps = 4/94 (4%)
Query 52 MSKEKYQKQWEAISHKIEKANSELLCLSYGSLVSQLLKDFEQVEAVNAQLEKMGYNIGVR 111
MSK K E + K++K N+EL L+YGS+V+QL KD E VN +L+KMGYNIG+R
Sbjct 1 MSKSKIG---EDVYKKVDKVNAELFVLTYGSIVAQLCKDM-NYEKVNEELDKMGYNIGIR 56
Query 112 LVDEFLAKANIGACESFRETAEVIAKLGLRMFLG 145
L+++FLAK C FRET E +AK+G ++FL
Sbjct 57 LIEDFLAKTEWPRCADFRETGETVAKVGFKVFLN 90
> CE00368
Length=181
Score = 84.3 bits (207), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 35/79 (44%), Positives = 58/79 (73%), Gaps = 1/79 (1%)
Query 69 EKANSELLCLSYGSLVSQLLKDFEQVEAVNAQLEKMGYNIGVRLVDEFLAK-ANIGACES 127
+K ++EL CL+YG++V+++LKD+E + V QL+KMG+N+G RL D+FLAK AN+ C
Sbjct 13 KKMSAELFCLTYGAMVTEMLKDYEDPKDVTIQLDKMGFNMGTRLADDFLAKNANVPRCVD 72
Query 128 FRETAEVIAKLGLRMFLGV 146
R+ A+V+ + + +LG+
Sbjct 73 TRQIADVLCRNAIPCYLGI 91
> YKR068c
Length=193
Score = 83.6 bits (205), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 38/86 (44%), Positives = 59/86 (68%), Gaps = 1/86 (1%)
Query 62 EAISHKIEKANSELLCLSYGSLVSQLLKDFEQ-VEAVNAQLEKMGYNIGVRLVDEFLAKA 120
E +K EK N+EL L+YGS+V+QL +D+E+ VN L MGYNIG RL+++FLA+
Sbjct 16 EIWKNKTEKINTELFTLTYGSIVAQLCQDYERDFNKVNDHLYSMGYNIGCRLIEDFLART 75
Query 121 NIGACESFRETAEVIAKLGLRMFLGV 146
+ CE+ +T+EV++K ++FL +
Sbjct 76 ALPRCENLVKTSEVLSKCAFKIFLNI 101
> 7294941
Length=143
Score = 63.9 bits (154), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 27/60 (45%), Positives = 43/60 (71%), Gaps = 1/60 (1%)
Query 87 LLKDFEQVEAVNAQLEKMGYNIGVRLVDEFLAKANIGACESFRETAEVIAKLGLRMFLGV 146
+L+DFE E VN QLE++GYN+G+RL+++FLA+ + C RETA+ I + R++L +
Sbjct 1 MLRDFENAEDVNKQLERIGYNMGMRLIEDFLARTSAPRCLEMRETADRIQQ-AFRIYLNI 59
> SPACUNK4.16c
Length=944
Score = 32.7 bits (73), Expect = 0.23, Method: Composition-based stats.
Identities = 26/80 (32%), Positives = 45/80 (56%), Gaps = 6/80 (7%)
Query 38 SAAAESARAAGFTTMSKEKYQKQWEAISHKIEKANSELLCLSYGSLVSQLLK---DFEQV 94
S AE+ R A TMS+E+ QK+ +A+ + I + ++ +++ SL+ + K D +++
Sbjct 606 SKTAEAFRTA--LTMSEEECQKRNKAMCNLILRHDAASWAVTFQSLIKESWKEQIDMQRI 663
Query 95 EAVNAQLEKMGY-NIGVRLV 113
A AQL K Y N RL+
Sbjct 664 PAFTAQLIKEPYQNAQKRLI 683
> At4g39660
Length=447
Score = 30.8 bits (68), Expect = 1.1, Method: Composition-based stats.
Identities = 24/83 (28%), Positives = 38/83 (45%), Gaps = 5/83 (6%)
Query 63 AISHKIEKANSELLCLSYGSLVSQLLKDFEQVEAVNAQLEKMGYNIGVRLV----DEFLA 118
A+ + I+K + C GS + Q LKD ++ + + G +G+ LV D+ A
Sbjct 330 AVLNVIDKEKRQEHCAEVGSHLIQRLKDVQKRHDIIGDVRGRGLMVGIELVSDRKDKTPA 389
Query 119 KANIGAC-ESFRETAEVIAKLGL 140
KA E RE ++ K GL
Sbjct 390 KAETSVLFEQLRELGILVGKGGL 412
> CE06498
Length=1307
Score = 30.0 bits (66), Expect = 1.4, Method: Composition-based stats.
Identities = 15/37 (40%), Positives = 21/37 (56%), Gaps = 0/37 (0%)
Query 54 KEKYQKQWEAISHKIEKANSELLCLSYGSLVSQLLKD 90
K K +W+AIS + E + ++CL SL LLKD
Sbjct 801 KNKTIDEWDAISPEAEHLDCIIMCLHCVSLAQSLLKD 837
> SPAC222.14c
Length=762
Score = 30.0 bits (66), Expect = 1.7, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 90 DFEQVEAVNAQLEKMGYNIGVRLVDEFLAKAN 121
+FE EAV +K+ +N+ L++EFLA N
Sbjct 462 EFEVSEAVEVAFQKLSHNVWDTLLNEFLAAQN 493
> CE13900
Length=881
Score = 29.6 bits (65), Expect = 2.1, Method: Composition-based stats.
Identities = 13/35 (37%), Positives = 20/35 (57%), Gaps = 1/35 (2%)
Query 53 SKEKYQKQWEAISHKIEKANSELLCLSYGSLVSQL 87
++EK QKQW ++H+ E N + SY + QL
Sbjct 573 TREKLQKQWRGVAHEYEHFNVTVF-QSYSFYIDQL 606
> HsM5031585
Length=748
Score = 28.9 bits (63), Expect = 3.3, Method: Composition-based stats.
Identities = 13/51 (25%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 50 TTMSKEKYQKQWEAISHKIEKANSELLCLSYGSLVSQLLKDFEQVEAVNAQ 100
TT ++ +KQ++ I H E +++L+ S G S ++F + +N +
Sbjct 485 TTPGAQEGKKQYKMICHVFESEDAQLIAQSIGQAFSVAYQEFLRANGINPE 535
> Hs9954875
Length=610
Score = 28.9 bits (63), Expect = 3.5, Method: Composition-based stats.
Identities = 16/52 (30%), Positives = 29/52 (55%), Gaps = 3/52 (5%)
Query 56 KYQKQWEAISHKIEKANSELLCLSYGSLVSQLLKDFEQVEAVNAQLEKMGYN 107
K +++WE + +EK +EL+CL +L + EQ Q+E++GY+
Sbjct 495 KQRQEWE-VKALLEKVPAELICLDPRALAEVDVISLEQ--GKKEQIERLGYD 543
> Hs22035550
Length=749
Score = 28.9 bits (63), Expect = 3.7, Method: Composition-based stats.
Identities = 13/51 (25%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 50 TTMSKEKYQKQWEAISHKIEKANSELLCLSYGSLVSQLLKDFEQVEAVNAQ 100
TT ++ +KQ++ I H E +++L+ S G S ++F + +N +
Sbjct 486 TTPGAQEGKKQYKMICHVFESEDAQLIAQSIGQAFSVAYQEFLRANGINPE 536
> Hs4501955
Length=1014
Score = 28.5 bits (62), Expect = 4.4, Method: Composition-based stats.
Identities = 24/92 (26%), Positives = 40/92 (43%), Gaps = 13/92 (14%)
Query 23 AASVATAAAESAAAESAAAESARAA-----GFTTMSKEKYQKQWEAISHKIEK------- 70
+ASVA S A+ AA S+ +A ++ K + + + IEK
Sbjct 362 SASVAATPPPSTASAPAAVNSSASADKPLSNMKILTLGKLSRNKDEVKAMIEKLGGKLTG 421
Query 71 -ANSELLCLSYGSLVSQLLKDFEQVEAVNAQL 101
AN LC+S V ++ K E+V+ N ++
Sbjct 422 TANKASLCISTKKEVEKMNKKMEEVKEANIRV 453
> Hs6912456
Length=580
Score = 27.7 bits (60), Expect = 7.8, Method: Composition-based stats.
Identities = 20/71 (28%), Positives = 30/71 (42%), Gaps = 0/71 (0%)
Query 20 VRVAASVATAAAESAAAESAAAESARAAGFTTMSKEKYQKQWEAISHKIEKANSELLCLS 79
V++ AA A + + A G T S + + WE I K +K SEL L+
Sbjct 414 VQLVKKFIKIAAHCKAQRNLNSFFAIVMGLNTASVSRLSQTWEKIPGKFKKLFSELESLT 473
Query 80 YGSLVSQLLKD 90
SL + +D
Sbjct 474 DPSLNHKAYRD 484
Lambda K H
0.317 0.126 0.343
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1748847648
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40