bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2651_orf1
Length=97
Score E
Sequences producing significant alignments: (Bits) Value
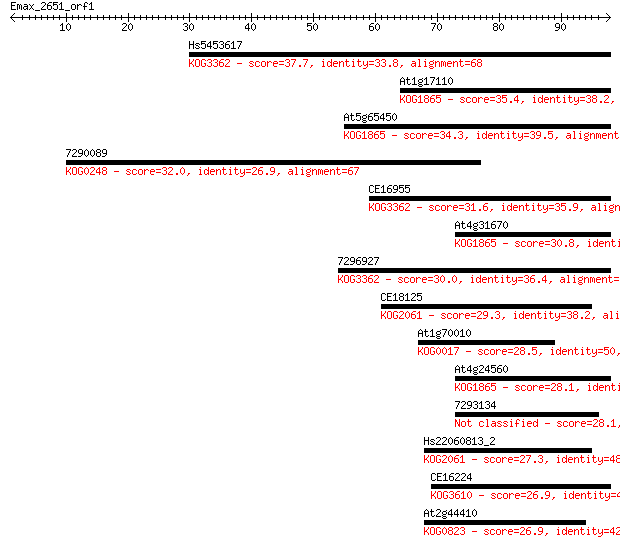
Hs5453617 37.7 0.005
At1g17110 35.4 0.025
At5g65450 34.3 0.056
7290089 32.0 0.30
CE16955 31.6 0.37
At4g31670 30.8 0.67
7296927 30.0 1.1
CE18125 29.3 1.9
At1g70010 28.5 3.1
At4g24560 28.1 3.8
7293134 28.1 3.8
Hs22060813_2 27.3 7.1
CE16224 26.9 8.4
At2g44410 26.9 9.4
> Hs5453617
Length=154
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 34/68 (50%), Gaps = 6/68 (8%)
Query 30 RERRNLGGWLQSRRSIPLELTVPGPSYMAMAAPPPLHLGGYHLCSVCLLPASYKCVRCRG 89
R R+N L+ + E GP+Y+ A PP C+VC P+ Y CV C G
Sbjct 79 RFRKNFQALLEEQNLSVAE----GPNYLTACAGPPSR-PQRPFCAVCGFPSPYTCVSC-G 132
Query 90 ALFCSIKC 97
A +C+++C
Sbjct 133 ARYCTVRC 140
> At1g17110
Length=891
Score = 35.4 bits (80), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 13/34 (38%), Positives = 20/34 (58%), Gaps = 0/34 (0%)
Query 64 PLHLGGYHLCSVCLLPASYKCVRCRGALFCSIKC 97
P++ H+C+ C PA +C RC+ +CS KC
Sbjct 121 PVNNNELHVCARCFGPAKTRCSRCKSVRYCSGKC 154
> At5g65450
Length=731
Score = 34.3 bits (77), Expect = 0.056, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 26/49 (53%), Gaps = 6/49 (12%)
Query 55 SYMAMAA-PPPLHLGG-----YHLCSVCLLPASYKCVRCRGALFCSIKC 97
SY+A P + GG + C+VCL P + +C +C+ +CS KC
Sbjct 33 SYLAEEVRPATVDYGGDSVSDVYRCAVCLYPTTTRCSQCKSVRYCSSKC 81
> 7290089
Length=1376
Score = 32.0 bits (71), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 18/67 (26%), Positives = 30/67 (44%), Gaps = 0/67 (0%)
Query 10 GGVHVNKISVNVVQTVSDVGRERRNLGGWLQSRRSIPLELTVPGPSYMAMAAPPPLHLGG 69
G +++ V V+ VSD E R Q+R ++ + GP+Y+ ++ P
Sbjct 798 GQINMRDARVEEVEHVSDSDSEEREDAAQDQARLTVAIYPAHQGPTYLILSGKPERDNWL 857
Query 70 YHLCSVC 76
YHL V
Sbjct 858 YHLTVVS 864
> CE16955
Length=263
Score = 31.6 bits (70), Expect = 0.37, Method: Compositional matrix adjust.
Identities = 14/39 (35%), Positives = 22/39 (56%), Gaps = 1/39 (2%)
Query 59 MAAPPPLHLGGYHLCSVCLLPASYKCVRCRGALFCSIKC 97
+++ PP C+VC + + Y C RC GA +CS+ C
Sbjct 212 LSSAPPSEKPARKFCAVCGIISKYCCTRC-GAKYCSLPC 249
> At4g31670
Length=631
Score = 30.8 bits (68), Expect = 0.67, Method: Composition-based stats.
Identities = 11/25 (44%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 73 CSVCLLPASYKCVRCRGALFCSIKC 97
CSVC ++ KC RC+ +CS +C
Sbjct 61 CSVCGNFSTKKCSRCKSVRYCSAEC 85
> 7296927
Length=151
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 23/44 (52%), Gaps = 2/44 (4%)
Query 54 PSYMAMAAPPPLHLGGYHLCSVCLLPASYKCVRCRGALFCSIKC 97
P+Y + AAP P H C+VC + Y C C G +C ++C
Sbjct 96 PNYESAAAPAP-QKPLRHFCAVCGNFSLYSCTAC-GTRYCCVRC 137
> CE18125
Length=386
Score = 29.3 bits (64), Expect = 1.9, Method: Composition-based stats.
Identities = 13/34 (38%), Positives = 18/34 (52%), Gaps = 0/34 (0%)
Query 61 APPPLHLGGYHLCSVCLLPASYKCVRCRGALFCS 94
A P G LC +C A+ KC +C+ A +CS
Sbjct 134 ADPRAPADGPGLCRICGCSAAKKCAKCQVARYCS 167
> At1g70010
Length=1315
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 11/22 (50%), Positives = 13/22 (59%), Gaps = 0/22 (0%)
Query 67 LGGYHLCSVCLLPASYKCVRCR 88
L G H VC LPA +C+ CR
Sbjct 824 LEGTHTWEVCSLPADKRCIGCR 845
> At4g24560
Length=1008
Score = 28.1 bits (61), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 11/25 (44%), Positives = 15/25 (60%), Gaps = 0/25 (0%)
Query 73 CSVCLLPASYKCVRCRGALFCSIKC 97
C VC A+ +C RC+ +CS KC
Sbjct 74 CPVCYCLATTRCSRCKAVRYCSGKC 98
> 7293134
Length=308
Score = 28.1 bits (61), Expect = 3.8, Method: Composition-based stats.
Identities = 9/23 (39%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 73 CSVCLLPASYKCVRCRGALFCSI 95
C++C A ++C C G LFC++
Sbjct 255 CNICNEDADFRCHGCGGELFCTL 277
> Hs22060813_2
Length=141
Score = 27.3 bits (59), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 14/27 (51%), Gaps = 0/27 (0%)
Query 68 GGYHLCSVCLLPASYKCVRCRGALFCS 94
G HLCSVC C RC A +CS
Sbjct 72 SGAHLCSVCGCLGPKTCSRCHKAYYCS 98
> CE16224
Length=1806
Score = 26.9 bits (58), Expect = 8.4, Method: Composition-based stats.
Identities = 12/29 (41%), Positives = 14/29 (48%), Gaps = 1/29 (3%)
Query 69 GYHLCSVCLLPASYKCVRCRGALFCSIKC 97
GY CS C+ + Y C C G CS C
Sbjct 620 GYGTCSSCM-SSEYNCAWCSGLHKCSNSC 647
> At2g44410
Length=404
Score = 26.9 bits (58), Expect = 9.4, Method: Composition-based stats.
Identities = 11/26 (42%), Positives = 15/26 (57%), Gaps = 0/26 (0%)
Query 68 GGYHLCSVCLLPASYKCVRCRGALFC 93
GG+ C++CL A + C G LFC
Sbjct 111 GGFFDCNICLEKAEDPILTCCGHLFC 136
Lambda K H
0.323 0.139 0.451
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1198045380
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40