bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2675_orf1
Length=142
Score E
Sequences producing significant alignments: (Bits) Value
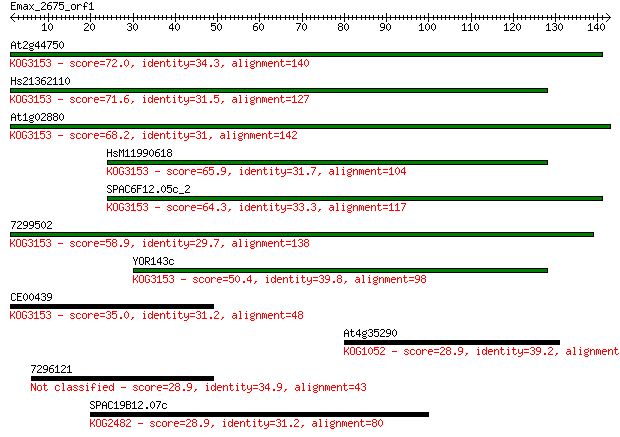
At2g44750 72.0 3e-13
Hs21362110 71.6 5e-13
At1g02880 68.2 5e-12
HsM11990618 65.9 2e-11
SPAC6F12.05c_2 64.3 6e-11
7299502 58.9 3e-09
YOR143c 50.4 9e-07
CE00439 35.0 0.047
At4g35290 28.9 3.4
7296121 28.9 3.5
SPAC19B12.07c 28.9 3.6
> At2g44750
Length=263
Score = 72.0 bits (175), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 48/143 (33%), Positives = 75/143 (52%), Gaps = 11/143 (7%)
Query 1 DQIHTDLEKAFAF-AASKFRLSHDDIVIIA-GAIGGRFDHSITAVSFLYKVHNASAETLP 58
DQ TDL+K ++ S + I+A GA+GGRFDH ++ LY+ +
Sbjct 108 DQDTTDLDKCISYIRHSTLNQESSRLQILATGALGGRFDHEAGNLNVLYRYPDT------ 161
Query 59 PKVILLGECNACLLLPEGD-NEVLLSDGVFSEACGLLPMAGEVRNVTTEGLKWNVKGEPL 117
+++LL + LLP+ +E+ + + CGL+P+ N TT GLKW++ +
Sbjct 162 -RIVLLSDDCLIQLLPKTHRHEIHIHSSLQGPHCGLIPIGTPSANTTTSGLKWDLSNTEM 220
Query 118 RVGGLISSSNIRIKVRIKIATSD 140
R GGLIS+SN+ +K I SD
Sbjct 221 RFGGLISTSNL-VKEEIITVESD 242
> Hs21362110
Length=243
Score = 71.6 bits (174), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 40/129 (31%), Positives = 66/129 (51%), Gaps = 6/129 (4%)
Query 1 DQIHTDLEKAFAFAASKFRLS--HDDIVIIAGAIGGRFDHSITAVSFLYKVHNASAETLP 58
DQ HTD K K D+++ G + GRFD + +V+ L++ + + P
Sbjct 95 DQDHTDFTKCLKMLQKKIEEKDLKVDVIVTLGGLAGRFDQIMASVNTLFQATHIT----P 150
Query 59 PKVILLGECNACLLLPEGDNEVLLSDGVFSEACGLLPMAGEVRNVTTEGLKWNVKGEPLR 118
+I++ E + LL G + + + G+ + CGL+P+ VTT GLKWN+ + L
Sbjct 151 FPIIIIQEESLIYLLQPGKHRLHVDTGMEGDWCGLIPVGQPCMQVTTTGLKWNLTNDVLA 210
Query 119 VGGLISSSN 127
G L+S+SN
Sbjct 211 FGTLVSTSN 219
> At1g02880
Length=277
Score = 68.2 bits (165), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 44/145 (30%), Positives = 74/145 (51%), Gaps = 10/145 (6%)
Query 1 DQIHTDLEKAFAF-AASKFRLSHDDIVIIA-GAIGGRFDHSITAVSFLYKVHNASAETLP 58
DQ TDL+K + S + I+A GA+GGRFDH ++ LY+ +
Sbjct 122 DQDTTDLDKCILYIRHSTLNQETSGLQILATGALGGRFDHEAGNLNVLYRYPDT------ 175
Query 59 PKVILLGECNACLLLPEGD-NEVLLSDGVFSEACGLLPMAGEVRNVTTEGLKWNVKGEPL 117
+++LL + LLP+ +E+ + + CGL+P+ TT GL+W++ +
Sbjct 176 -RIVLLSDDCLIQLLPKTHRHEIHIQSSLEGPHCGLIPIGTPSAKTTTSGLQWDLSNTEM 234
Query 118 RVGGLISSSNIRIKVRIKIATSDPL 142
R GGLIS+SN+ + +I + + L
Sbjct 235 RFGGLISTSNLVKEEKITVESDSDL 259
> HsM11990618
Length=137
Score = 65.9 bits (159), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 33/104 (31%), Positives = 59/104 (56%), Gaps = 4/104 (3%)
Query 24 DIVIIAGAIGGRFDHSITAVSFLYKVHNASAETLPPKVILLGECNACLLLPEGDNEVLLS 83
D+++ G + GRFD + +V+ L++ + + P +I++ E + LL G + + +
Sbjct 14 DVIVTLGGLAGRFDQIMASVNTLFQATHIT----PFPIIIIQEESLIYLLQPGKHRLHVD 69
Query 84 DGVFSEACGLLPMAGEVRNVTTEGLKWNVKGEPLRVGGLISSSN 127
G+ + CGL+P+ VTT GLKWN+ + L G L+S+SN
Sbjct 70 TGMEGDWCGLIPVGQPCMQVTTTGLKWNLTNDVLAFGTLVSTSN 113
> SPAC6F12.05c_2
Length=233
Score = 64.3 bits (155), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 39/117 (33%), Positives = 60/117 (51%), Gaps = 4/117 (3%)
Query 24 DIVIIAGAIGGRFDHSITAVSFLYKVHNASAETLPPKVILLGECNACLLLPEGDNEVLLS 83
D + + +GGR DH+I ++ L+ + S + +V LL E N LL G N V
Sbjct 111 DTIFVLCGMGGRVDHAIGNLNHLFWAASISEKN---EVFLLTELNVSTLLQPGINHVDCH 167
Query 84 DGVFSEACGLLPMAGEVRNVTTEGLKWNVKGEPLRVGGLISSSNIRIKVRIKIATSD 140
D + CGLLP+ V T GL+WN++ + GGL+SS N+ K + I ++
Sbjct 168 DNI-GLHCGLLPVGQSVYVKKTSGLEWNIEDRICQFGGLVSSCNVVTKATVTIEVNN 223
> 7299502
Length=332
Score = 58.9 bits (141), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 41/139 (29%), Positives = 60/139 (43%), Gaps = 5/139 (3%)
Query 1 DQIHTDLEKAFAFAASKFRLSHDDIVIIAGAIGGRFDHSITAVSFLYKVHNASAETLPPK 60
DQ TD KA A V++ GR D + ++ LYK +
Sbjct 175 DQDATDFTKAMAVLQPVMTQRKIQDVVVFHDTSGRLDQVMANLNTLYKSQKDNCNVF--- 231
Query 61 VILLGECNACLLLPEGDNEVLLSDGVFSEA-CGLLPMAGEVRNVTTEGLKWNVKGEPLRV 119
+L G+ LL P + D V S+ C L+P+ NVTT GLKWN+ L
Sbjct 232 -LLSGDSVTWLLRPGKHTIQVPVDLVTSQRWCSLMPVGSSAHNVTTTGLKWNLYHAQLEF 290
Query 120 GGLISSSNIRIKVRIKIAT 138
GG++S+SN +++ T
Sbjct 291 GGMVSTSNTYATEFVQVET 309
> YOR143c
Length=319
Score = 50.4 bits (119), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 39/106 (36%), Positives = 54/106 (50%), Gaps = 15/106 (14%)
Query 30 GAIGGRFDHSITAVSFLYKV-HNASAETL----PPKVILLGECNACLLL--PEGDNEVLL 82
G IGGRFD ++ +++ LY + NAS L P +I L + N L+ P+ N +
Sbjct 190 GGIGGRFDQTVHSITQLYTLSENASYFKLCYMTPTDLIFLIKKNGTLIEYDPQFRNTCIG 249
Query 83 SDGVFSEACGLLPMAGEVRNVTTEGLKWNVKGEPLR-VGGLISSSN 127
+ CGLLP+ T GLKW+VK P V G +SSSN
Sbjct 250 N-------CGLLPIGEATLVKETRGLKWDVKNWPTSVVTGRVSSSN 288
> CE00439
Length=195
Score = 35.0 bits (79), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 15/50 (30%), Positives = 30/50 (60%), Gaps = 2/50 (4%)
Query 1 DQIHTDLEKAFAFAASKFRLSHDDI--VIIAGAIGGRFDHSITAVSFLYK 48
DQ +TDL K+ + + L+ + +++ G + GRFDH+++ +S L +
Sbjct 89 DQDYTDLSKSVQWCLEQKTLTSWEFENIVVLGGLNGRFDHTMSTLSSLIR 138
> At4g35290
Length=925
Score = 28.9 bits (63), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 30/64 (46%), Gaps = 17/64 (26%)
Query 80 VLLSDGVFSEACGLLP-------------MAGEVRNVTTEGLKWNVKGEPLRVGGLISSS 126
VL+ DG+ SE GL P + GEV N+ + + +V +P +GG S
Sbjct 13 VLIGDGMISEGAGLRPRYVDVGAIFSLGTLQGEVTNIAMKAAEEDVNSDPSFLGG----S 68
Query 127 NIRI 130
+RI
Sbjct 69 KLRI 72
> 7296121
Length=531
Score = 28.9 bits (63), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 21/43 (48%), Gaps = 0/43 (0%)
Query 6 DLEKAFAFAASKFRLSHDDIVIIAGAIGGRFDHSITAVSFLYK 48
DLEKA F A + L DD+ I G D SI ++ + +
Sbjct 114 DLEKALDFRAGRLSLGDDDVSIELGMGTSCIDSSINVINVILQ 156
> SPAC19B12.07c
Length=319
Score = 28.9 bits (63), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 37/81 (45%), Gaps = 9/81 (11%)
Query 20 LSHDDIVIIAGAIGGRFD-HSITAVSFLYKVHNASAETLPPKVILLGECNACLLLPEGDN 78
+S DD +II F SI+ +S L +HN+S +TL NA L E N
Sbjct 1 MSSDDAIIIRCPFDCEFAGDSISFISHLTTIHNSSVQTLAK--------NAYLCASEIAN 52
Query 79 EVLLSDGVFSEACGLLPMAGE 99
E+ + + + LL +A E
Sbjct 53 ELNQNPNLKDQEILLLQIAQE 73
Lambda K H
0.320 0.138 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1603110344
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40