bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Emax_2714_orf1
Length=70
Score E
Sequences producing significant alignments: (Bits) Value
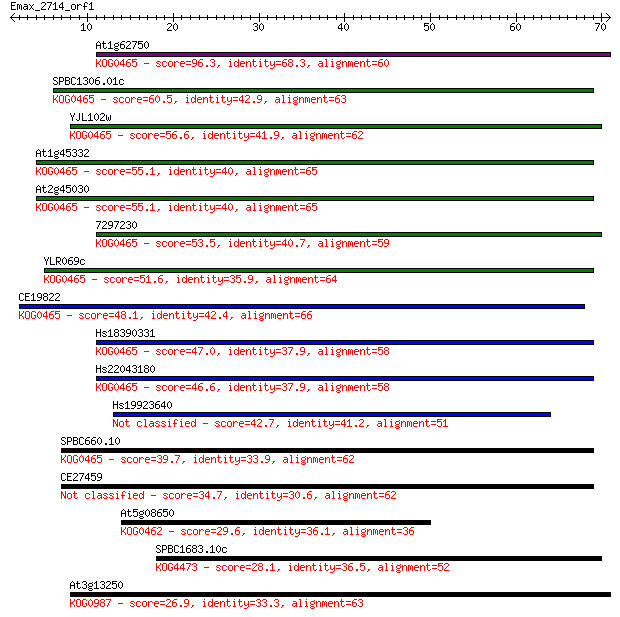
At1g62750 96.3 1e-20
SPBC1306.01c 60.5 7e-10
YJL102w 56.6 1e-08
At1g45332 55.1 3e-08
At2g45030 55.1 3e-08
7297230 53.5 9e-08
YLR069c 51.6 3e-07
CE19822 48.1 4e-06
Hs18390331 47.0 1e-05
Hs22043180 46.6 1e-05
Hs19923640 42.7 2e-04
SPBC660.10 39.7 0.001
CE27459 34.7 0.040
At5g08650 29.6 1.3
SPBC1683.10c 28.1 4.1
At3g13250 26.9 8.4
> At1g62750
Length=783
Score = 96.3 bits (238), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 41/60 (68%), Positives = 51/60 (85%), Gaps = 0/60 (0%)
Query 11 DDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMHANSR 70
DD +P A LAFKI +DPFVG LTF+RVY+G + +GSYVLNA+K +ER+GRLL+MHANSR
Sbjct 391 DDDEPFAGLAFKIMSDPFVGSLTFVRVYSGKISAGSYVLNANKGKKERIGRLLEMHANSR 450
> SPBC1306.01c
Length=558
Score = 60.5 bits (145), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 27/63 (42%), Positives = 44/63 (69%), Gaps = 1/63 (1%)
Query 6 VELMADDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQM 65
V L+ +KPL ALAFK+ F G+LT++R+Y G+L+ G+Y+ N + + +V RL++M
Sbjct 157 VSLVPSSEKPLVALAFKLEEGRF-GQLTYLRIYQGTLKRGNYIYNVNSTKKIKVSRLVRM 215
Query 66 HAN 68
H+N
Sbjct 216 HSN 218
> YJL102w
Length=819
Score = 56.6 bits (135), Expect = 1e-08, Method: Composition-based stats.
Identities = 26/62 (41%), Positives = 42/62 (67%), Gaps = 0/62 (0%)
Query 8 LMADDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMHA 67
L+ ++K ALAFK+ TDP G+ FIR+Y+G+L SG+ V N++ + ++G+LL HA
Sbjct 350 LVNNNKNLCIALAFKVITDPIRGKQIFIRIYSGTLNSGNTVYNSTTGEKFKLGKLLIPHA 409
Query 68 NS 69
+
Sbjct 410 GT 411
> At1g45332
Length=754
Score = 55.1 bits (131), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 41/65 (63%), Gaps = 1/65 (1%)
Query 4 DEVELMADDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLL 63
+ V L PL ALAFK+ F G+LT++RVY G ++ G +++N + R +V RL+
Sbjct 354 ERVTLTGSPDGPLVALAFKLEEGRF-GQLTYLRVYEGVIKKGDFIINVNTGKRIKVPRLV 412
Query 64 QMHAN 68
+MH+N
Sbjct 413 RMHSN 417
> At2g45030
Length=754
Score = 55.1 bits (131), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 41/65 (63%), Gaps = 1/65 (1%)
Query 4 DEVELMADDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLL 63
+ V L PL ALAFK+ F G+LT++RVY G ++ G +++N + R +V RL+
Sbjct 354 ERVTLTGSPDGPLVALAFKLEEGRF-GQLTYLRVYEGVIKKGDFIINVNTGKRIKVPRLV 412
Query 64 QMHAN 68
+MH+N
Sbjct 413 RMHSN 417
> 7297230
Length=729
Score = 53.5 bits (127), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 24/59 (40%), Positives = 35/59 (59%), Gaps = 1/59 (1%)
Query 11 DDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMHANS 69
D K P LAFK+ F G+LT++R Y G L G + NA N + R+ RL+++H+N
Sbjct 326 DGKDPFVGLAFKLEAGRF-GQLTYLRCYQGVLRKGDNIFNARTNKKVRIARLVRLHSNQ 383
> YLR069c
Length=761
Score = 51.6 bits (122), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 40/64 (62%), Gaps = 1/64 (1%)
Query 5 EVELMADDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQ 64
+V L+ ++P LAFK+ + G+LT++RVY G L G+Y+ N + +V RL++
Sbjct 364 KVNLVPAVQQPFVGLAFKLEEGKY-GQLTYVRVYQGRLRKGNYITNVKTGKKVKVARLVR 422
Query 65 MHAN 68
MH++
Sbjct 423 MHSS 426
> CE19822
Length=750
Score = 48.1 bits (113), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 28/70 (40%), Positives = 38/70 (54%), Gaps = 5/70 (7%)
Query 2 TGDEVELMADDK----KPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRE 57
TGDE ++ K KP LAFK+ + G+LT+ RVY G L G V + +
Sbjct 332 TGDEKGIILSPKRNNDKPFVGLAFKLEAGKY-GQLTYFRVYQGQLSKGDTVYASRDGRKV 390
Query 58 RVGRLLQMHA 67
RV RL++MHA
Sbjct 391 RVQRLVRMHA 400
> Hs18390331
Length=751
Score = 47.0 bits (110), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 33/58 (56%), Gaps = 1/58 (1%)
Query 11 DDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMHAN 68
D+ P LAFK+ F G+LT++R Y G L+ G + N + R+ RL +MHA+
Sbjct 348 DNSHPFVGLAFKLEVGRF-GQLTYVRSYQGELKKGDTIYNTRTRKKVRLQRLARMHAD 404
> Hs22043180
Length=485
Score = 46.6 bits (109), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 22/58 (37%), Positives = 33/58 (56%), Gaps = 1/58 (1%)
Query 11 DDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMHAN 68
D+ P LAFK+ F G+LT++R Y G L+ G + N + R+ RL +MHA+
Sbjct 82 DNSHPFVGLAFKLEVGRF-GQLTYVRSYQGELKKGDTIYNTRTRKKVRLQRLARMHAD 138
> Hs19923640
Length=779
Score = 42.7 bits (99), Expect = 2e-04, Method: Composition-based stats.
Identities = 21/51 (41%), Positives = 31/51 (60%), Gaps = 0/51 (0%)
Query 13 KKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLL 63
K L ALAFK+ D G L F+R+Y+G+++ + N + N ER+ RLL
Sbjct 364 KDDLCALAFKVLHDKQRGPLVFMRIYSGTIKPQLAIHNINGNCTERISRLL 414
> SPBC660.10
Length=813
Score = 39.7 bits (91), Expect = 0.001, Method: Composition-based stats.
Identities = 21/62 (33%), Positives = 33/62 (53%), Gaps = 0/62 (0%)
Query 7 ELMADDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMH 66
++++ DK+PL A FK+ G LT++RV G+L G + N ER RL +
Sbjct 329 KIISLDKRPLLAKIFKVIHHASRGILTYVRVNEGTLSRGMMMFNPRTKKSERAIRLYNVF 388
Query 67 AN 68
A+
Sbjct 389 AD 390
> CE27459
Length=689
Score = 34.7 bits (78), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 32/62 (51%), Gaps = 0/62 (0%)
Query 7 ELMADDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMH 66
EL K + I D G+L+++R+YTGSL + S + N S+ + E +L +
Sbjct 264 ELTCASKIATISCGSAITHDKRRGQLSYMRIYTGSLHNNSTIFNTSQMTSEGPLKLFTPY 323
Query 67 AN 68
A+
Sbjct 324 AD 325
> At5g08650
Length=675
Score = 29.6 bits (65), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 13/36 (36%), Positives = 19/36 (52%), Gaps = 0/36 (0%)
Query 14 KPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVL 49
KPL AL F DP+ G + + RV G ++ G +
Sbjct 264 KPLRALIFDSYYDPYRGVIVYFRVIDGKVKKGDRIF 299
> SPBC1683.10c
Length=242
Score = 28.1 bits (61), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 19/58 (32%), Positives = 29/58 (50%), Gaps = 6/58 (10%)
Query 18 ALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNA------SKNSRERVGRLLQMHANS 69
+L IAT F+G L F+ + G + +L A + + VGRLL++HA S
Sbjct 180 SLVLPIATMFFLGMLGFVGAHIGGAKRVRAILRAVVLGLLAMAATALVGRLLEIHALS 237
> At3g13250
Length=1419
Score = 26.9 bits (58), Expect = 8.4, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 27/63 (42%), Gaps = 1/63 (1%)
Query 8 LMADDKKPLAALAFKIATDPFVGRLTFIRVYTGSLESGSYVLNASKNSRERVGRLLQMHA 67
L ADD+ + A FKI D + LT R G + Y + K LL M A
Sbjct 409 LTADDRPDIVARIFKIKLDSLMKDLT-ERHLLGKTVASMYTVEFQKRGLPHAHILLFMAA 467
Query 68 NSR 70
NS+
Sbjct 468 NSK 470
Lambda K H
0.317 0.132 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1197219688
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40